Sequential Non-Saccharomyces and Saccharomyces cerevisiae Fermentations to Reduce the Alcohol Content in Wine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains: Purity and Identity Control
2.2. Laboratory-Scale Fermentations
2.3. Analitycal Determination of Wines
2.4. Statistical Treatment of Data
3. Results
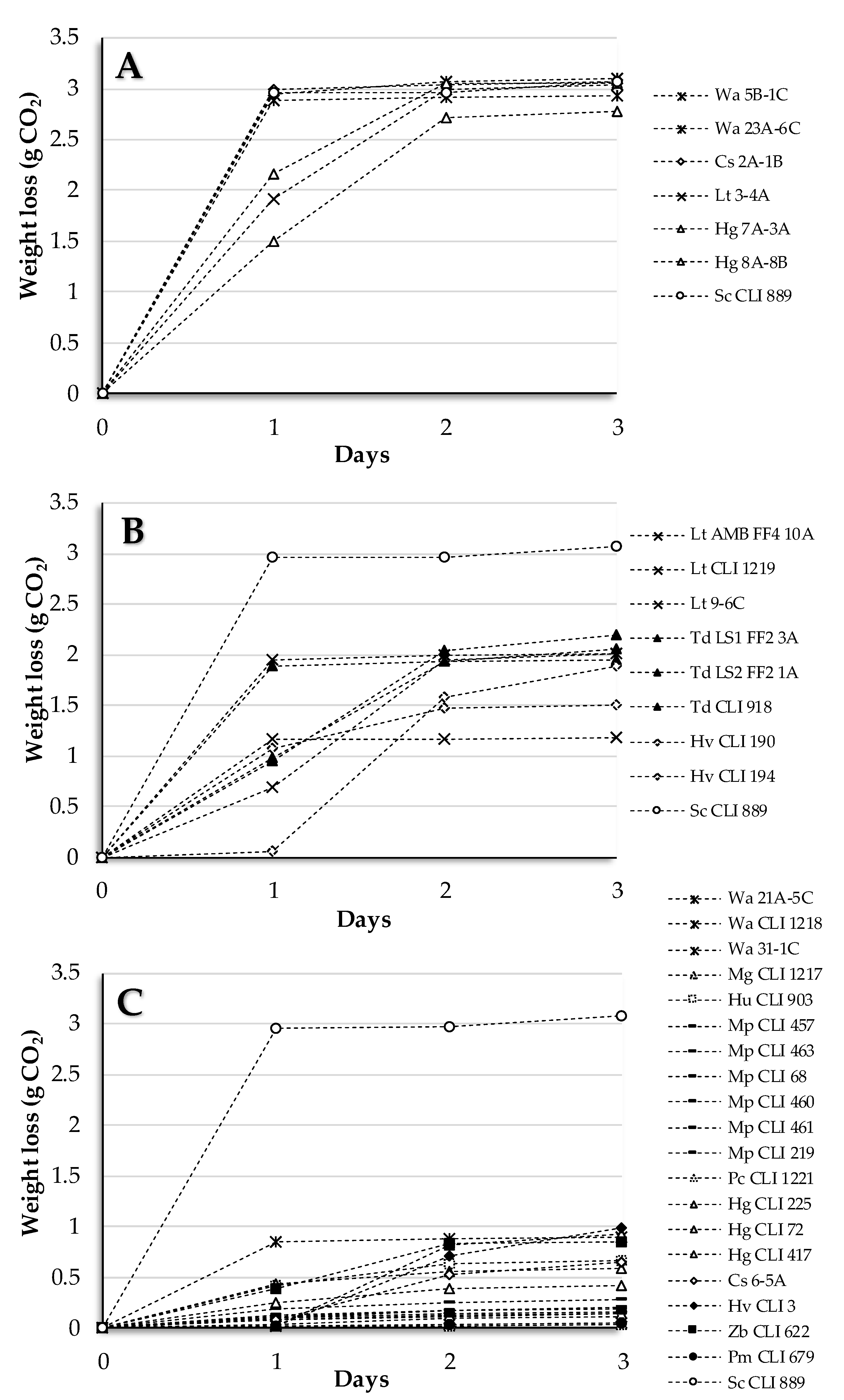
3.1. Section I: Pure Culture of Non-Saccharomyces Strains
3.2. Section II: Sequential Culture of Non-Saccharomyces/S. cerevisiae Strains
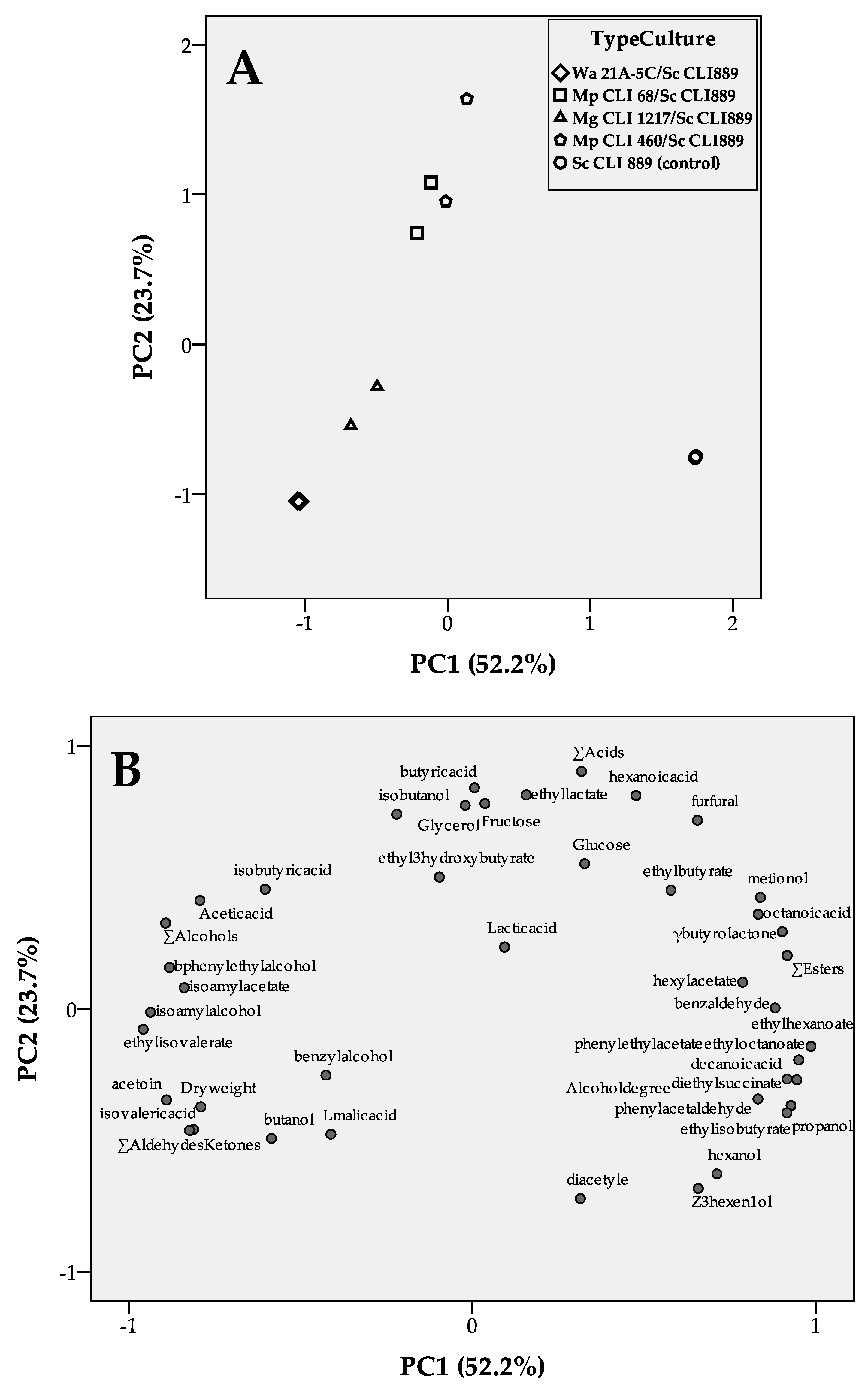
3.3. Yeast Strain Sequential Combinations Selected as Low-Ethanol Producers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mira de Orduña, R. Climate change associated effects on grape and wine quality and production. Food Res. Int. 2010, 43, 1844–1855. [Google Scholar] [CrossRef]
- Gonzalez, R.; Quirós, M.; Morales, P. Yeast respiration of sugars by non-Saccharomyces yeast species: A promising and barely explored approach to lowering alcohol content of wines. Trends Food Sci. Technol. 2013, 29, 55–61. [Google Scholar] [CrossRef]
- Liguori, L.; Russo, P.; Albanese, D.; Di Matteo, M. Production of low-alcohol beverages: Current status and perspectives. In Food Processing for Increased Quality and Consumption; Grumezescu, A., Holban, A.M., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 347–382. ISBN 9780128114476. [Google Scholar]
- Stockley, C.S.; Varela, C.; Coulter, A.; Dry, P.R.; Francis, I.L.; Muhlack, R.; Pretorius, I.S. Controlling the Highs and the Lows of Alcohol in Wine; Nova Science Publishers, Inc.: New York, NY, USA, 2012; ISBN 9781614706359. [Google Scholar]
- Varela, C.; Dry, P.R.; Kutyna, D.R.; Francis, I.L.; Henschke, P.A.; Curtin, C.D.; Chambers, P.J. Strategies for reducing alcohol concentration in wine. Aust. J. Grape Wine Res. 2015, 21, 670–679. [Google Scholar] [CrossRef]
- Fischer, U.; Noble, A. The effect of ethanol, catechin concentration, and pH on sourness and bitterness of wine. Am. J. Enol. Vitic. 1994, 45, 6–10. [Google Scholar]
- Wilkinson, K.L.; Jiranck, V. Wine of reduced alcohol content: Consumer and society demand vs. industry willingness and ability to deliver. In 1st International Symposium Alcohol Level Reduction in Wine; Institut des Sciences de la Vigne et du Vin: Villenave d’Ornon CEDEX, France, 2013; pp. 98–104. [Google Scholar]
- Stockwell, T.; Zhao, J.; Panwar, S.; Roemer, A.; Naimi, T.; Chikritzhs, T. Do “moderate” drinkers have reduced mortality risk? A systematic review and meta-analysis of alcohol consumption and all-cause mortality. J. Stud. Alcohol Drugs 2016, 77, 185–198. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Vandenberg, B.; Hollingsworth, B. Minimum pricing of alcohol versus volumetric taxation: Which policy will reduce heavy consumption without adversely affecting light and moderate consumers? PLoS ONE 2014, 9, e80936. [Google Scholar] [CrossRef]
- Ciani, M.; Morales, P.; Comitini, F.; Tronchoni, J.; Canonico, L.; Curiel, J.A.; Oro, L.; Rodrigues, A.J.; Gonzalez, R. Non-conventional yeast species for lowering ethanol content of wines. Front. Microbiol. 2016, 7, 642. [Google Scholar] [CrossRef] [Green Version]
- Rolle, L.; Englezos, V.; Torchio, F.; Cravero, F.; Río Segade, S.; Rantsiou, K.; Giacosa, S.; Gambuti, A.; Gerbi, V.; Cocolin, L. Alcohol reduction in red wines by technological and microbiological approaches: A comparative study. Aust. J. Grape Wine Res. 2017, 24, 62–74. [Google Scholar] [CrossRef]
- Tilloy, V.; Ortiz-Julien, A.; Dequin, S. Reduction of ethanol yield and improvement of glycerol formation by adaptive evolution of the wine yeast Saccharomyces cerevisiae under hyperosmotic conditions. Appl. Environ. Microbiol. 2014, 80, 2623–2632. [Google Scholar] [CrossRef] [Green Version]
- Fleet, G.H.; Heard, G.M. Yeast growth during fermentation. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Chur, Switzerland, 1993; pp. 27–54. [Google Scholar]
- Andorrà, I.; Berradre, M.; Rozès, N.; Mas, A.; Guillamón, J.M.; Esteve-Zarzoso, B. Effect of pure and mixed cultures of the main wine yeast species on grape must fermentations. Eur. Food Res. Technol. 2010, 231, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Cominiti, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- García, M.; Arroyo, T.; Crespo, J.; Cabellos, J.M.; Esteve-Zarzoso, B. Use of native non-Saccharomyces strain: A new strategy in D.O. “Vinos de Madrid” (Spain) wines elaboration. Eur. J. Food Sci. Technol. 2017, 5, 1–31. [Google Scholar]
- Dutraive, O.; Benito, S.; Fritsch, S.; Beisert, B.; Patz, C.; Rauhut, D. Effect of sequential inoculation with non-Saccharomyces and Saccharomyces yeasts on Riesling wine chemical composition. Fermentation 2019, 5, 79. [Google Scholar] [CrossRef] [Green Version]
- Cuello, R.A.; Flores Montero, K.J.; Mercado, L.A.; Combina, M.; Ciklic, I.F. Construction of low-ethanol–wine yeasts through partial deletion of the Saccharomyces cerevisiae PDC2 gene. AMB Express 2017, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Contreras, A.; Hidalgo, C.; Schmidt, S.; Henschke, P.A.; Curtin, C.; Varela, C. The application of non-Saccharomyces yeast in fermentations with limited aeration as a strategy for the production of wine with reduced alcohol content. Int. J. Food Microbiol. 2015, 205, 7–15. [Google Scholar] [CrossRef]
- Ciani, M.; Beco, L.; Comitini, F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006, 108, 239–245. [Google Scholar] [CrossRef]
- Magyar, I.; Tóth, T. Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 94–100. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 2014, 80, 1670–1678. [Google Scholar] [CrossRef] [Green Version]
- Morales, P.; Rojas, V.; Quirós, M.; Gonzalez, R. The impact of oxygen on the final alcohol content of wine fermented by a mixed starter culture. Appl. Microbiol. Biotechnol. 2015, 99, 3993–4003. [Google Scholar] [CrossRef] [Green Version]
- Röcker, J.; Strub, S.; Ebert, K.; Grossmann, M. Usage of different aerobic non-Saccharomyces yeasts and experimental conditions as a tool for reducing the potential ethanol content in wines. Eur. Food Res. Technol. 2016, 242, 2051–2070. [Google Scholar] [CrossRef]
- Canonico, L.; Comitini, F.; Oro, L.; Ciani, M. Sequential fermentation with selected immobilized non-Saccharomyces yeast for reduction of ethanol content in wine. Front. Microbiol. 2016, 7, 278. [Google Scholar] [CrossRef] [Green Version]
- Varela, C.; Barker, A.; Tran, T.; Borneman, A.; Curtin, C. Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. Int. J. Food Microbiol. 2017, 252, 1–9. [Google Scholar] [CrossRef]
- Canonico, L.; Comitini, F.; Ciani, M. Metschnikowia pulcherrima selected strain for ethanol reduction in wine: Influence of cell immobilization and aeration condition. Foods 2019, 8, 378. [Google Scholar] [CrossRef] [Green Version]
- Aplin, J.J.; White, K.P.; Edwards, C.G. Growth and metabolism of non-Saccharomyces yeasts isolated from Washington state vineyards in media and high sugar grape musts. Food Microbiol. 2019, 77, 158–165. [Google Scholar] [CrossRef]
- Puškaš, V.S.; Miljić, U.D.; Djuran, J.J.; Vučurović, V.M. The aptitude of commercial yeast strains for lowering the ethanol content of wine. Food Sci. Nutr. 2020, 8, 278. [Google Scholar] [CrossRef]
- Quirós, M.; Rojas, V.; Gonzalez, R.; Morales, P. Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 2014, 181, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Cordero-Bueso, G.; Esteve-Zarzoso, B.; Cabellos, J.M.; Gil-Díaz, M.; Arroyo, T. Biotechnological potential of non-Saccharomyces yeasts isolated during spontaneous fermentations of Malvar (Vitis vinifera cv. L.). Eur. Food Res. Technol. 2013, 236, 193–207. [Google Scholar] [CrossRef]
- Gil, M.; Cabellos, J.M.; Arroyo, T.; Prodanov, M. Characterization of the volatile fraction of young wines from the Denomination of Origin “Vinos de Madrid” (Spain). Anal. Chim. Acta 2006, 563, 145–153. [Google Scholar] [CrossRef]
- Tello, J.; Cordero-Bueso, G.; Aporta, I.; Cabellos, J.M.; Arroyo, T. Genetic diversity in commercial wineries: Effects of the farming system and vinification management on wine yeasts. J. Appl. Microbiol. 2012, 112, 302–315. [Google Scholar] [CrossRef]
- Cordero-Bueso, G.; Esteve-Zarzoso, B.; Gil-Díaz, M.; García, M.; Cabellos, J.; Arroyo, T. Improvement of Malvar wine quality by use of locally-selected Saccharomyces cerevisiae strains. Fermentation 2016, 2, 7. [Google Scholar] [CrossRef] [Green Version]
- Merloni, E.; Camanzi, L.; Mulazzani, L.; Malorgio, G. Adaptive capacity to climate change in the wine industry: A Bayesian Network approach. Wine Econ. Policy 2018, 7, 165–177. [Google Scholar] [CrossRef]
- Arroyo, T.; Cordero, G.; Serrano, A.; Valero, E. β-Glucosidase production by non-Saccharomyces yeasts isolated from vineyard. In Expression of Multidisciplinary Flavour Science; Blank, I., Wüst, M., Yeretzian, C., Eds.; Institute of Chemistry and Biological Chemistry: Winterthur, Switzerland, 2010; pp. 359–362. [Google Scholar]
- Arroyo, T. Estudio de la Influencia de Diferentes Tratamientos Enológicos en la Evolución de la Microbiota y en la Calidad de los vinos Elaborados con la Variedad “Airén”, en la D.O. “Vinos de Madrid”. Ph.D. Thesis, University of Alcalá, Madrid, Spain, 2000. [Google Scholar]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordero-Bueso, G.; Arroyo, T.; Serrano, A.; Tello, J.; Aporta, I.; Vélez, M.D.; Valero, E. Influence of the farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 2011, 145, 132–139. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Esteve-Zarzoso, B.; Crespo, J.; Cabellos, J.M.; Arroyo, T. Yeast monitoring of wine mixed or sequential fermentations made by native strains from D.O. “Vinos de Madrid” using real-time quantitative PCR. Front. Microbiol. 2017, 8, 2520. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- García, M.; Greetham, D.; Wimalasena, T.T.; Phister, T.G.; Cabellos, J.M.; Arroyo, T. The phenotypic characterization of yeast strains to stresses inherent to wine fermentation in warm climates. J. Appl. Microbiol. 2016, 121, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, M.; Apolinar-Valiente, R.; Williams, P.; Esteve-Zarzoso, B.; Arroyo, T.; Crespo, J.; Doco, T. Polysaccharides and oligosaccharides produced on Malvar wines elaborated with Torulaspora delbrueckii CLI 918 and Saccharomyces cerevisiae CLI 889 native yeasts from D.O. “Vinos de Madrid.”. J. Agric. Food Chem. 2017, 65, 6656–6664. [Google Scholar] [CrossRef]
- Ortega, C.; López, R.; Cacho, J.; Ferreira, V. Fast analysis of important wine volatile compounds-Development and validation of a new method based on gas chromatographic-flame ionisation detection analysis of dichloromethane microextracts. J. Chromatogr. A 2001, 923, 205–214. [Google Scholar] [CrossRef]
- Balboa-Lagunero, T.; Arroyo, T.; Cabellos, J.M.; Aznar, M. Yeast selection as a tool for reducing key oxidation notes in organic wines. Food Res. Int. 2013, 53, 252–259. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine. In Food Microbiology: Fundamentals and Frontiers; Doyle, M.P., Beuchat, L.R., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 863–890. [Google Scholar]
- Ciani, M.; Canonico, L.; Oro, L.; Comitini, F. Sequential fermentation using non-Saccharomyces yeasts for the reduction of alcohol content in wine. BIOWeb Conf. 2014, 3, 02015. [Google Scholar] [CrossRef]
- Loira, I.; Vejarano, R.; Bañuelos, M.A.; Morata, A.; Tesfaye, W.; Uthurry, C.; Villa, A.; Cintora, I.; Suárez-Lepe, J.A. Influence of sequential fermentation with Torulaspora delbrueckii and Saccharomyces cerevisiae on wine quality. LWT Food Sci. Technol. 2014, 59, 915–922. [Google Scholar] [CrossRef] [Green Version]
- Vilela, A. Modulating wine pleasantness throughout wine-yeast co-inoculation or sequential inoculation. Fermentation 2020, 6, 22. [Google Scholar] [CrossRef] [Green Version]
- Ciani, M.; Ferraro, L. Enhanced glycerol content in wines made with immobilized Candida stellata cells. Appl. Environ. Microbiol. 1996, 62, 128–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erten, H.; Campbell, I. The production of low-alcohol wines by aerobic yeasts. J. Inst. Brew. 2001, 107, 207–215. [Google Scholar] [CrossRef]
- Rodrigues, A.J.; Raimbourg, T.; Gonzalez, R.; Morales, P. Environmental factors influencing the efficacy of different yeast strains for alcohol level reduction in wine by respiration. LWT Food Sci. Technol. 2016, 65, 1038–1043. [Google Scholar] [CrossRef]
- Ferraro, L.; Fatichenti, F.; Ciani, M. Pilot scale vinification process using immobilized Candida stellata cells and Saccharomyces cerevisiae. Process Biochem. 2000, 35, 1125–1129. [Google Scholar] [CrossRef]
- Canonico, L.; Solomon, M.; Comitini, F.; Ciani, M.; Varela, C. Volatile profile of reduced alcohol wines fermented with selected non-Saccharomyces yeasts under different aeration conditions. Food Microbiol. 2019, 84, 103247. [Google Scholar] [CrossRef]
- Contreras, A.; Curtin, C.; Varela, C. Yeast population dynamics reveal a potential “collaboration” between Metschnikowia pulcherrima and Saccharomyces uvarum for the production of reduced alcohol wines during Shiraz fermentation. Appl. Microbiol. Biotechnol. 2014, 99, 1885–1895. [Google Scholar] [CrossRef]
- Mestre Furlani, M.V.; Maturano, Y.P.; Combina, M.; Mercado, L.A.; Toro, M.E.; Vazquez, F. Selection of non-Saccharomyces yeasts to be used in grape musts with high alcoholic potential: A strategy to obtain wines with reduced ethanol content. FEMS Yeast Res. 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remize, F.; Roustan, J.L.; Sablayrolles, J.M.; Barre, P.; Dequin, S. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 1999, 65, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Eglinton, J.M.; Heinrich, A.J.; Pollnitz, A.P.; Langridge, P.; Henschke, P.A.; De Barros Lopes, M. Decreasing acetic acid accumulation by a glycerol overproducing strain of Saccharomyces cerevisiae by deleting the ALD6 aldehyde dehydrogenase gene. Yeast 2002, 19, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Sadoudi, M.; Rousseaux, S.; David, V.; Alexandre, H.; Tourdot-Maréchal, R. Metschnikowia pulcherrima influences the expression of genes involved in PDH bypass and glyceropyruvic fermentation in Saccharomyces cerevisiae. Front. Microbiol. 2017, 8, 1137. [Google Scholar] [CrossRef]
- Varela, C.; Kutyna, D.R.; Solomon, M.R.; Black, C.A.; Borneman, A.; Henschke, P.A.; Pretorius, I.S.; Chambers, P.J. Evaluation of gene modification strategies for the development of low-alcohol-wine yeasts. Appl. Environ. Microbiol. 2012, 78, 6068–6077. [Google Scholar] [CrossRef] [PubMed]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Beltran, G.; Esteve-Zarzoso, B.; Rozès, N.; Mas, A.; Guillamón, J.M. Influence of the timing of nitrogen additions during synthetic grape must fermentations on fermentation kinetics and nitrogen consumption. J. Agric. Food Chem. 2005, 53, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int. J. Food Microbiol. 2012, 157, 245–250. [Google Scholar] [CrossRef]
- Vilanova, M.; Ugliano, M.; Varela, C.; Siebert, T.; Pretorius, I.S.; Henschke, P.A. Assimilable nitrogen utilisation and production of volatile and non-volatile compounds in chemically defined medium by Saccharomyces cerevisiae wine yeasts. Appl. Microbiol. Biotechnol. 2007, 77, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Styger, G.; Prior, B.; Bauer, F.F. Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 2011, 38, 1145–1159. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial contribution to wine aroma and its intended use for wine quality improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef] [Green Version]
- Rojas, V.; Gil, J.V.; Piñaga, F.; Manzanares, P. Studies on acetate ester production by non-Saccharomyces wine yeasts. Int. J. Food Microbiol. 2001, 70, 283–289. [Google Scholar] [CrossRef]
- Izquierdo Cañas, P.M.; Palacios García, A.T.; García Romero, E. Enhancement of flavour properties in wines using sequential inoculations of non-Saccharomyces (Hansenula and Torulaspora) and Saccharomyces yeast starter. Vitis 2011, 50, 177–182. [Google Scholar]
- Domizio, P.; Romani, C.; Lencioni, L.; Comitini, F.; Gobbi, M.; Mannazzu, I.; Ciani, M. Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 2011, 147, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo Cañas, P.M.; García-Romero, E.; Heras Manso, J.M.; Fernández-González, M. Influence of sequential inoculation of Wickerhamomyces anomalus and Saccharomyces cerevisiae in the quality of red wines. Eur. Food Res. Technol. 2014, 239, 279–286. [Google Scholar] [CrossRef]
- Sáez, J.S.; Lopes, C.A.; Kirs, V.C.; Sangorrín, M.P. Enhanced volatile phenols in wine fermented with Saccharomyces cerevisiae and spoiled with Pichia guilliermondii and Dekkera bruxellensis. Lett. Appl. Microbiol. 2010, 51, 170–176. [Google Scholar] [CrossRef]
- Suárez, R.; Suárez-Lepe, J.A.; Morata, A.; Calderón, F. The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera: A review. Food Chem. 2007, 102, 10–21. [Google Scholar] [CrossRef]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef]
- González-Royo, E.; Pascual, O.; Kontoudakis, N.; Esteruelas, M.; Esteve-Zarzoso, B.; Mas, A.; Canals, J.M.; Zamora, F. Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 2015, 240, 999–1012. [Google Scholar] [CrossRef]
- Gobert, A.; Tourdot-Maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexandre, H. Non-Saccharomyces yeasts nitrogen source preferences: Impact on sequential fermentation and wine volatile compounds profile. Front. Microbiol. 2017, 8, 2175. [Google Scholar] [CrossRef] [Green Version]
- Seguinot, P.; Ortiz-Julien, A.; Camarasa, C. Impact of nutrient availability on the fermentation and production of aroma compounds under sequential inoculation with M. pulcherrima and S. cerevisiae. Front. Microbiol. 2020, 11, 305. [Google Scholar] [CrossRef]
- Rodríguez, M.E.; Lopes, C.A.; Barbagelata, R.J.; Barda, N.B.; Caballero, A.C. Influence of Candida pulcherrima Patagonian strain on alcoholic fermentation behaviour and wine aroma. Int. J. Food Microbiol. 2010, 138, 19–25. [Google Scholar] [CrossRef]
- Varela, C.; Sengler, F.; Solomon, M.; Curtin, C. Volatile flavour profile of reduced alcohol wines fermented with the non-conventional yeast species Metschnikowia pulcherrima and Saccharomyces uvarum. Food Chem. 2016, 209, 57–64. [Google Scholar] [CrossRef]
- Ruiz, J.; Belda, I.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Santos, A.; Benito, S. Analytical impact of Metschnikowia pulcherrima in the volatile profile of Verdejo white wines. Appl. Microbiol. Biotechnol. 2018, 102, 8501–8509. [Google Scholar] [CrossRef] [Green Version]
- Lambrechts, M.G.; Pretorius, I.S. Yeast and its importance to wine aroma-A review. South African J. Enol. Vitic. 2000, 21, 97–129. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Zhang, Z. Volatile compounds of young wines from Cabernet Sauvignon, Cabernet Gernischet and Chardonnay varieties grown in the Loess Plateau region of China. Molecules 2010, 15, 9184–9196. [Google Scholar] [CrossRef] [Green Version]
| Species Name | Strain Code | Year of Isolation | Origin 1 | References 2 |
|---|---|---|---|---|
| Wickerhamomyces anomalus | CLI 1218 | 2007 | Malvar a | [31,36] |
| 31-1C | 2006 | Garnacha c | This study | |
| 21A-5C | 2007 | Garnacha c | [36] | |
| 23A-6C | 2007 | Garnacha c | [36] | |
| 5B-1C | 2008 | Garnacha c | This study | |
| Candida stellata | 6-5A | 2006 | Shiraz c | [36] |
| 2A-1B | 2007 | Shiraz c | This study | |
| Hanseniaspora valbyensis | CLI 194 | 1993 | Garnacha a | [36] |
| CLI 190 | 1993 | Garnacha a | [36] | |
| Hanseniaspora guilliermondii | CLI 417 | 1995 | Malvar a | This study |
| 7A-3A | 2007 | Garnacha c | This study | |
| 8A-8B | 2007 | Garnacha c | This study | |
| CLI 225 | 1994 | Tempranillo a | [36,42] | |
| CLI 72 | 1993 | Garnacha a | [36] | |
| Hanseniaspora uvarum | CLI 903 | 1993 | Airén b | [36,42] |
| Hanseniaspora vineae | CLI 3 | 1993 | Tempranillo a | [36] |
| Torulaspora delbrueckii | LS1 FF2 3A | 2009 | Garnacha a | [33] |
| LS2 FF2 1A | 2009 | Garnacha a | [33] | |
| CLI 918 | 2006 | Malvar a | [16,40,42,43] | |
| Metschnikowia pulcherrima | CLI 68 | 1993 | Garnacha a | [36] |
| CLI 457 | 1995 | Malvar a | [16,36,40,42] | |
| CLI 463 | 1995 | Malvar a | This study | |
| CLI 219 | 1994 | Malvar a | [36,42] | |
| CLI 460 | 1995 | Malvar a | [36,42] | |
| CLI 461 | 1995 | Malvar a | This study | |
| Lachancea thermotolerans | AMB FF4 10A | 2009 | Malvar a | [33] |
| 3-4A | 2006 | Shiraz c | [36] | |
| 9-6C | 2006 | Malvar a | [16,40,42] | |
| CLI 1219 | 2007 | Malvar a | [31,42] | |
| Pichia membranifaciens | CLI 679 | 2006 | Malvar a | [31,42] |
| Meyerozyma guilliermondii | CLI 1217 | 2006 | Malvar a | [31,42] |
| Priceomyces carsonii | CLI 1221 | 2006 | Malvar a | [31,42] |
| Zygosaccharomyces bailii | CLI 622 | 2009 | Malvar a | [31,42] |
| Saccharomyces cerevisiae | CLI 889 | 2000 | Airén a | [16,34,37,40,42,43] |
| Parameters | Yeast Culture | ||||
|---|---|---|---|---|---|
| Wa 21A-5C(S) | Mp CLI 68(S) | Mg CLI 1217(S) | Mp CLI 460(S) | Sc CLI 889(P) | |
| Malic acid (g/L) | 0.66 ± 0.12 a | 0.60 ± 0.02 a | 0.64 ± 0.02 a | 0.45 ± 0.09 a | 0.55 ± 0.02 a |
| Lactic acid (g/L) | 2.10 ± 0.26 a | 2.48 ± 0.30 a | 2.50 ± 0.23 a | 2.22 ± 0.53 a | 2.31 ± 0.02 a |
| Acetic acid (g/L) | 0.77 ± 0.08 a | 0.86 ± 0.01 ab | 0.78 ± 0.01 ab | 0.71 ± 0.16 a | 0.43 ± 0.00 ac |
| Glucose (g/L) | 2.45 ± 0.30 a | 2.80 ± 0.54 a | 2.04 ± 0.39 a | 2.97 ± 0.53 a | 2.70 ± 0.01 a |
| Fructose (g/L) | 1.06 ± 0.10 a | 2.72 ± 0.60 b | 0.69 ± 0.04 ac | 1.93 ± 0.29 ab | 1.01 ± 0.02 a |
| Glycerol (g/L) | 7.83 ± 0.31 a | 8.32 ± 0.10 a | 7.06 ± 0.61 ab | 9.30 ± 0.90 ac | 7.60 ± 0.02 a |
| Alcohol degree (%) | 12.05 ± 0.12 a | 11.75 ± 0.05 a | 11.77 ± 0.32 a | 12.16 ± 0.26 a | 13.00 ± 0.01 b |
| Dry weight (mg) | 4.35 ± 0.07 a | 3.73 ± 0.08 a | 4.77 ± 0.57 ab | 3.28 ± 0.61 a | 2.95 ± 0.01 ac |
| Compound | Wa 21A-5C(S) | Mp CLI 68(S) | Mg CLI 1217(S) | Mp CLI 460(S) | Sc CLI 889(P) |
|---|---|---|---|---|---|
| 1-Propanol | n.q. | n.q. | n.q. | n.q. | 3.69 ± 0.13 a |
| 1-Butanol | 1.81 ± 0.14 a | 0.50 ± 0.05 b | 0.46 ± 0.05 b | 0.48 ± 0.12 b | 0.40 ± 0.10 b |
| Isobutanol | 31.96 ± 1.31 a | 33.51 ± 4.11 a | 30.51 ± 1.28 a | 49.46 ± 0.24 b | 26.30 ± 0.95 a |
| Isoamyl alcohol | 118.13 ± 1.88 a | 114.56 ± 5.92 ab | 122.03 ± 0.33 a | 106.47 ± 0.53 b | 91.37 ± 3.14 c |
| (Z)-3-Hexen-1-ol | 0.12 ± 0.00 a | 0.04 ± 0.00 b | 0.05 ± 0.00 c | 0.04 ± 0.00 b | 0.20 ± 0.10 d |
| 1-Hexanol | 0.49 ± 0.00 a | 0.26 ± 0.00 b | 0.23 ± 0.03 b | 0.22 ± 0.00 b | 0.88 ± 0.05 c |
| Metionol | 0.09 ± 0.00 a | 0.37 ± 0.00 ab | 0.32 ± 0.00 ab | 0.53 ± 0.20 b | 0.61 ± 0.10 b |
| Benzyl alcohol | 0.17 ± 0.00 a | 0.19 ± 0.00 b | 0.27 ± 0.00 c | 0.13 ± 0.00 d | 0.15 ± 0.06 e |
| β-Phenylethyl alcohol | 27.55 ± 0.05 a | 21.27 ± 2.02 b | 18.93 ± 1.04 b | 23.03 ± 1.40 ab | 10.53 ± 0.29 c |
| ∑ Alcohols | 181.48 ± 3.37a | 171.86 ± 11.99a | 173.94 ± 0.01a | 181.51 ± 1.83a | 134.13 ± 4.92b |
| Ethyl butyrate | 0.21 ± 0.03 a | 0.30 ± 0.01 a | 0.30 ± 0.04 a | 0.29 ± 0.03 a | 0.31 ± 0.05 a |
| Ethyl isovalerate | 1.35 ± 0.07 a | 0.81 ± 0.01 b | 0.98 ± 0.10 b | 0.90 ± 0.06 b | 0.28 ± 0.05 c |
| Ethyl isobutyrate | n.q. | n.q. | n.q. | n.q. | 2.60 ± 0.37 a |
| Isoamyl acetate | 2.07 ± 0.02 a | 1.97 ± 0.04 a | 2.80 ± 0.00 b | 1.96 ± 0.17 a | 0.99 ± 0.05 c |
| Ethyl hexanoate | 0.03 ± 0.00 a | 0.21 ± 0.00 b | 0.13 ± 0.03 ab | 0.20 ± 0.05 b | 0.70 ± 0.06 c |
| Ethyl-3-hydroxybutyrate | 0.16 ± 0.00 a | 0.57 ± 0.00 b | 0.68 ± 0.00 c | 0.47 ± 0.00 d | 0.32 ± 0.06 e |
| Hexyl acetate | 0.05 ± 0.00 a | 0.05 ± 0.00 a | 0.06 ± 0.00 a | 0.07 ± 0.00 a | 0.07 ± 0.05 a |
| 2-Phenylethyl acetate | 0.31 ± 0.01 a | 0.28 ± 0.01 a | 0.24 ± 0.09 a | 0.39 ± 0.06 a | 0.76 ± 0.09 b |
| Diethyl succinate | n.q. | 0.09 ± 0.00 a | 0.05 ± 0.00 a | 0.26 ± 0.08 b | 6.57 ± 0.13 c |
| Ethyl octanoate | 0.06 ± 0.00 a | 0.17 ± 0.00 b | 0.21 ± 0.00 b | 0.18 ± 0.04 b | 0.51 ± 0.06 c |
| Ethyl lactate | 1.71 ± 0.56 a | 8.23 ± 1.24 b | 1.73 ± 0.39 a | 5.93 ± 1.27 bc | 3.32 ± 0.11 ac |
| ∑ Esters | 5.98 ± 0.63a | 12.68 ± 1.19b | 7.19 ± 0.45a | 10.63 ± 1.14b | 16.43 ± 1.08c |
| Isobutyric acid | 4.86 ± 0.05 a | 4.99 ± 0.34 a | 3.25 ± 0.07 b | 4.62 ± 0.04 a | 2.89 ± 0.05 b |
| Butyric acid | 0.22 ± 0.00 a | 0.29 ± 0.01 a | 0.25 ± 0.00 a | 0.40 ± 0.14 a | 0.23 ± 0.06 a |
| Isovaleric acid | 1.82 ± 0.03 a | 1.25 ± 0.04 b | 1.29 ± 0.10 b | 0.76 ± 0.01 c | 0.74 ± 0.06 c |
| Hexanoic acid | 0.90 ± 0.01 a | 2.97 ± 0.65 abc | 2.03 ± 0.60 ac | 5.01 ± 0.76 b | 3.11 ± 0.34 bc |
| Octanoic acid | 0.41 ± 0.02 a | 1.88 ± 0.09 b | 1.26 ± 0.16 c | 1.45 ± 0.15 c | 2.18 ± 0.05 b |
| Decanoic acid | 0.09 ± 0.00 a | 0.18 ± 0.06 a | 0.07 ± 0.00 a | 0.14 ± 0.05 a | 0.73 ± 0.09 b |
| ∑ Acids | 8.30 ± 0.11a | 11.57 ± 0.13bc | 8.15 ± 0.60a | 12.37 ± 1.14c | 9.88 ± 0.65ab |
| Diacetyle | 0.58 ± 0.01 a | 0.46 ± 0.17 a | 0.51 ± 0.02 a | 0.48 ± 0.04 a | 0.63 ± 0.09 a |
| Furfural | n.q. | 0.07 ± 0.00 a | 0.04 ± 0.00 a | 0.08 ± 0.00 a | 0.07 ± 0.05 a |
| Benzaldehyde | 0.01 ± 0.00 a | 0.04 ± 0.00 ab | 0.06 ± 0.00 b | 0.05 ± 0.01 b | 0.09 ± 0.05 c |
| Phenylacetaldehyde | n.q. | n.q. | n.q. | n.q. | 0.52 ± 0.05 a |
| Acetoin | 5.19 ± 0.46 a | 1.48 ± 0.01 b | 3.71 ± 0.00 c | 2.30 ± 0.43 b | 0.20 ± 0.10 d |
| ∑ Aldehydes/Ketones | 5.78 ± 0.47a | 2.04 ± 0.15b | 4.32 ± 0.02c | 2.91 ± 0.45bd | 1.51 ± 0.34be |
| γ-Butyrolactone | 0.98 ± 0.00 a | 6.78 ± 0.13 b | 1.64 ± 0.00 c | 5.41 ± 0.07 d | 9.40 ± 0.10 e |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García, M.; Esteve-Zarzoso, B.; Cabellos, J.M.; Arroyo, T. Sequential Non-Saccharomyces and Saccharomyces cerevisiae Fermentations to Reduce the Alcohol Content in Wine. Fermentation 2020, 6, 60. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation6020060
García M, Esteve-Zarzoso B, Cabellos JM, Arroyo T. Sequential Non-Saccharomyces and Saccharomyces cerevisiae Fermentations to Reduce the Alcohol Content in Wine. Fermentation. 2020; 6(2):60. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation6020060
Chicago/Turabian StyleGarcía, Margarita, Braulio Esteve-Zarzoso, Juan Mariano Cabellos, and Teresa Arroyo. 2020. "Sequential Non-Saccharomyces and Saccharomyces cerevisiae Fermentations to Reduce the Alcohol Content in Wine" Fermentation 6, no. 2: 60. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation6020060