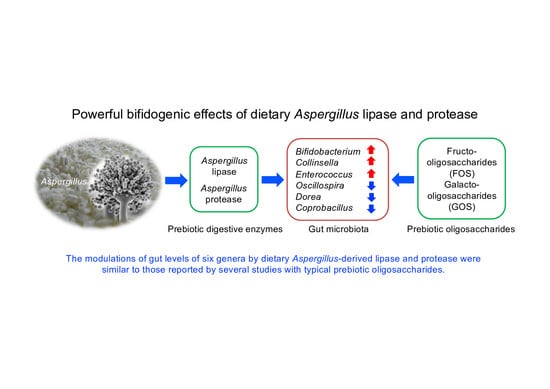
Supplemental Aspergillus Lipase and Protease Preparations Display Powerful Bifidogenic Effects and Modulate the Gut Microbiota Community of Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Sample Collection
2.3. 16S rRNA Gene-Based Microbiome Analysis
2.4. Bioinformatics Analysis
2.5. Analysis of Cecal Organic Acids
2.6. Data Analysis
2.7. Evaluation of the Risk of Bias in the Methodology
3. Results
3.1. Food intake, Body Weight, and Cecal Content Weight
3.2. Cecal Microbiota
3.3. Cecal Organic Acids
4. Discussion
4.1. Bifidobacterium
4.2. Collinsella
4.3. Enterococcus
4.4. Oscillospira, Dorea, and Coprobacillus
4.5. Parabacteroides and Allobaculum
4.6. Bacterial Diversity
4.7. Organic Acids
4.8. Limitations of this Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board and Statement
Data Availability Statement
Conflicts of Interest
References
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conlon, M.A.; Bird, A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef]
- Patel, S.; Goyal, A. The current trends and future perspectives of prebiotics research: A review. 3 Biotech 2012, 2, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.L.; Lin, T.L.; Chang, C.J.; Wu, T.R.; Lai, W.F.; Lu, C.C. Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 2019, 26, 3. [Google Scholar] [CrossRef]
- Stavropoulou, E.; Bezirtzoglou, E. Probiotics in Medicine: A Long Debate. Front. Immunol. 2020, 11, 2192. [Google Scholar] [CrossRef] [PubMed]
- Ichishima, E. Enzymology of the national microorganisms of Japan in a historical context. J. Biochem. Biotechnol. 2019, 2, 11–18. [Google Scholar]
- Takenaka, S.; Nakabayashi, R.; Ogawa, C.; Kimura, Y.; Yokota, S.; Doi, M. Characterization of surface Aspergillus community involved in traditional fermentation and ripening of katsuobushi. Int. J. Food Microbiol. 2020, 327, 108654. [Google Scholar] [CrossRef]
- Ianiro, G.; Pecere, S.; Giorgio, V.; Gasbarrini, A.; Cammarota, G. Digestive Enzyme Supplementation in Gastrointestinal Diseases. Curr. Drug Metabol. 2016, 17, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Sitanggang, N.V.; Kato, N.; Inoue, J.; Murakami, T.; Watanabe, T.; Iguchi, T.; Okazaki, Y. Beneficial effects of protease preparations derived from Aspergillus on the colonic luminal environment in rats consuming a high-fat diet. Biomed. Rep. 2015, 3, 715–720. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Iwamoto, A.; Kumrungsee, T.; Okazaki, Y.; Kuroda, M.; Yamaguchi, S.; Katoa, N. Consumption of an acid protease derived from Aspergillus oryzae causes bifidogenic effect in rats. Nutr. Res. 2017, 44, 60–66. [Google Scholar] [CrossRef]
- Yang, Y.; Kumrungsee, T.; Kuroda, M.; Yamaguchi, S.; Kato, N. Feeding Aspergillus protease preparation combined with adequate protein diet to rats increases levels of cecum gut-protective amino acids, partially linked to Bifidobacterium and Lactobacillus. Biosci. Biotech. Biochem. 2019, 83, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Van Hul, M.; Karnik, K.; Canene-Adams, K.; De Souza, M.; Abbeele, P.V.D.; Marzorati, M.; Delzenne, N.M.; Everard, A.; Cani, P.D. Comparison of the effects of soluble corn fiber and fructooligosaccharides on metabolism, inflammation, and gut microbiome of high-fat diet-fed mice. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E779–E791. [Google Scholar] [CrossRef]
- Ishii, C.; Nakanishi, Y.; Murakami, S.; Nozu, R.; Ueno, M.; Hioki, K.; Aw, W.; Hirayama, A.; Soga, T.; Ito, M.; et al. A metabologenomic approach reveals changes in the intestinal environment of mice fed on American diet. Int. J. Mol. Sci. 2018, 19, 4079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Zhu, M. A sensitive GC/MS detection method for analyzing microbial metabolites short chain fatty acids in fecal and serum samples. Talanta 2019, 196, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Hooijmans, C.R.; Rovers, M.M.; BM de Vries, R.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Meth. 2014, 14, 43. [Google Scholar] [CrossRef] [Green Version]
- Patterson, E.; Ryan, P.M.; Wiley, N.; Carafa, I.; Sherwin, E.; Moloney, G.; Franciosi, E.; Mandal, R.; Wishart, D.S.; Tuohy, K.; et al. Gamma-aminobutyric acid-producing lactobacilli positively affect metabolism and depressive-like behaviour in a mouse model of metabolic syndrome. Sci. Rep. 2019, 9, 16323. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Johnson, C.; Luk, B.; Uribe, G.; Fu, Y.; Oezguen, N.; Matsunami, R.K.; Lugo, M.; Major, A.; Mori-Akiyama, Y.; et al. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol. Motil. 2017, 29, e12904. [Google Scholar] [CrossRef]
- Basak, S.; Duttaroy, A.K. Conjugated Linoleic Acid and Its Beneficial Effects in Obesity, Cardiovascular Disease, and Cancer. Nutrients 2020, 12, 1913. [Google Scholar] [CrossRef]
- Carafa, I.; Masuero, D.; Vrhovsek, U.; Bittante, G.; Franciosi, E.; Tuohy, K.M. Production of conjugated linoleic acid (CLA): Effect of inulin on microbial composition and CLA concentration in a human intestinal model. Proc. Nutr. Soc. 2020, 79 (OCE2), E628. [Google Scholar] [CrossRef]
- Klancic, T.; Laforest-Lapointe, I.; Choo, A.; Nettleton, J.E.; Chleilat, F.; Noye Tuplin, E.W.; Alukic, E.; Cho, N.A.; Nicolucci, A.C.; Arrieta, M.; et al. Prebiotic Oligofructose Prevents Antibiotic-Induced Obesity Risk and Improves Metabolic and Gut Microbiota Profiles in Rat Dams and Offspring. Mol. Nutr. Food Res. 2020, 64, e2000288. [Google Scholar] [CrossRef]
- Dewulf, E.M.; Cani, P.D.; Claus, S.P.; Fuentes, S.; Puylaert, P.G.B.; Neyrinck, A.M.; Bindels, L.B.; de Vos, W.M.; Gibson, G.R.; Thissen, J.; et al. Insight into the prebiotic concept: Lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013, 62, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Kassinen, A.; Krogius-Kurikka, L.; Mäkivuokko, H.; Rinttilä, T.; Paulin, L.; Corander, J.; Malinen, E.; Apajalahti, J.; Palva, A. The Fecal Microbiota of Irritable Bowel Syndrome Patients Differs Significantly From That of Healthy Subjects. Gastroenterology 2007, 133, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Saalman, R.; Alderberth, I.; Wold, A.; Sjoberg, F. Use of Collinsella for Treatment of Inflammatory Bowel Disease. U.S. Patent No. 2018/0028579 A1, 1 February 2018. [Google Scholar]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, V.; Lambeth, S.M.; Carson, T.; Lowe, J.; Ramaraj, T.; Leff, J.W.; Luo, L.; Bell, J.C.; Shah, V.O. Composition Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. J. Diabetes Obes. 2015, 2, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns–An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Habermann, W.; Zimmermann, K.; Skarabis, H.; Kunze, R.; Rusch, V. The effect of a bacterial immunostimulant (human Enterococcus faecalis bacteria) on the occurrence of relapse in patients with chronic bronchtis. Arzneimittelforschung 2001, 51, 931–937. [Google Scholar]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef] [Green Version]
- Mao, B.Y.; Li, D.Y.; Zhao, J.X.; Liu, X.M.; Gu, Z.N.; Chen, Y.Q.; Zhang, H.; Chen, W. Metagenomic insights into the effects of fructo-oligosaccharides (FOS) on the composition of fecal microbiota in mice. J. Agric. Food Chem. 2015, 63, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; He, Y.; Li, H.; Yu, D.; Na, L.; Sun, T.; Zhang, D.; Shi, X.; Xia, Y.; Jiang, T.; et al. Effects of prebiotics on immunologic indicators and intestinal microbiota structure in perioperative colorectal cancer patients. Nutrition 2019, 61, 132–142. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol. Spectrum 2014, 5, 5–6. [Google Scholar]
- Lo Presti, A.; Zorzi, F.; Del Chierico, F.; Altomare, A.; Cocca, S.; Avola, A.; de Biasio, F.; Russo, A.; Cella, E.; Reddel, S.; et al. Fecal and Mucosal Microbiota Profiling in Irritable Bowel Syndrome and Inflammatory Bowel Disease. Front. Microbiol. 2019, 10, 1655. [Google Scholar] [CrossRef] [Green Version]
- Fei, Y.; Wang, Y.; Pang, Y.; Wang, W.; Zhu, D.; Xie, M.; Lan, S.; Wang, Z. Xylooligosaccharide Modulates Gut Microbiota and Alleviates Colonic Inflammation Caused by High Fat Diet Induced Obesity. Front. Physiol. 2020, 10, 1601. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pan, M.; Li, D.; Yin, Y.; Jiang, T.; Fang, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Metagenomic insights into the effects of oligosaccharides on the microbial composition of cecal contents in constipated mice. J. Funct. Foods 2017, 38, 486–496. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Barcenas-Walls, J.R.; Suchodolski, J.S.; Steiner, J.M. Molecular assessment of the fecal microbiota in healthy cats and dogs before and during supplementation with fructo-oligosaccharides (FOS) and inulin using high-throughput 454-pyrosequencing. PeerJ 2017, 5, e3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, C.; Cross, T.-W.L.; Devendran, S.; Neumer, F.; Theis, S.; Ridlon, J.M.; Suchodolski, J.S.; de Godoy, M.R.C.; Swanson, K.S. Effects of prebiotic inulin-type fructans on blood metabolite and hormone concentrations and faecal microbiota and metabolites in overweight dogs. Br. J. Nutr. 2018, 120, 711–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, A.; Lazarevic, V.; Gaïa, N.; Johansson, M.; Ståhlman, M.; Bäckhed, F.; Delzenne, N.; Schrenzel, J.; Francois, P.; Cani, P. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014, 8, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Audebert, C.; Even, G.; Cian, A.; Loywick, A.; Merlin, S.; Viscogliosi, E.; Chabe, M.; Blastocystis Investigation Group. Colonization with the Enteric Protozoa Blastocystis is Associated with Increased Diversity of Human Gut Bacterial Microbiota. Sci. Rep. 2016, 6, 25255. [Google Scholar] [CrossRef]
- Sjöberg, F.; Barkman, C.; Nookaew, I.; Östman, S.; Adlerberth, I.; Saalman, R.; Wold, A.E. Low-complexity microbiota in the duodenum of children with newly diagnosed ulcerative colitis. PLoS ONE 2017, 12, e0186178. [Google Scholar] [CrossRef] [Green Version]
- Shade, A. Diversity is the question, not the answer. ISME J. 2017, 11, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groussard, C.; Morel, I.; Chevanne, M.; Monnier, M.; Cillard, J.; Delamarche, A. Free radical scavenging and antioxidant effects of lactate ion: An in vitro study. J. Appl. Physiol. 2000, 89, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, I.; Prasad, P.D.; Thangaraju, M.; Manicassamy, S. Lactate-Dependent Regulation of Immune Responses by Dendritic Cells and Macrophages. Front. Immunol. 2021, 12, 691134. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Kumrungsee, T.; Kato, N.; Fukuda, S.; Kuroda, M.; Yamaguchi, S. Supplemental Aspergillus Lipase and Protease Preparations Display Powerful Bifidogenic Effects and Modulate the Gut Microbiota Community of Rats. Fermentation 2021, 7, 294. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation7040294
Yang Y, Kumrungsee T, Kato N, Fukuda S, Kuroda M, Yamaguchi S. Supplemental Aspergillus Lipase and Protease Preparations Display Powerful Bifidogenic Effects and Modulate the Gut Microbiota Community of Rats. Fermentation. 2021; 7(4):294. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation7040294
Chicago/Turabian StyleYang, Yongshou, Thanutchaporn Kumrungsee, Norihisa Kato, Shinji Fukuda, Manabu Kuroda, and Shotaro Yamaguchi. 2021. "Supplemental Aspergillus Lipase and Protease Preparations Display Powerful Bifidogenic Effects and Modulate the Gut Microbiota Community of Rats" Fermentation 7, no. 4: 294. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation7040294