Microbial Communities in Underground Gas Reservoirs Offer Promising Biotechnological Potential
Abstract
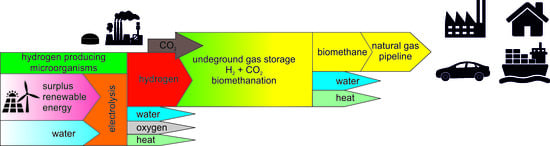
:1. Introduction
Methanogenesis
2. Materials and Methods
2.1. Locality and Geological Preconditions
2.2. Sampling
2.3. Physical-Chemical Parameters and Groundwater Chemical Composition
2.4. Degas Analysis
2.4.1. Isotopic Determination
2.4.2. Gas Chromatography
2.5. Microscopy
2.6. Molecular Biological Methods
2.6.1. DNA Isolation
2.6.2. Quantitave PCR
2.6.3. Illumina—Next Generation Sequencing Method
3. Results
3.1. Physicochemical Parameters and Water Chemistry
Characteristics of Groundwater
3.2. Isotopic Determination
3.3. Microscopy
3.4. Quantitative Analyses of Methanogens (qPCR)
3.5. Metagenomic Analyses of UGS Archaeal Community
3.5.1. Next-Generation Sequencing (NGS) Analysis
3.5.2. Biodiversity of Microbial Communities
3.5.3. Metabolism Prediction—FAPROTAX
4. Discussion
4.1. Lobodice
4.2. Dolní Dunajovice
4.3. Tvrdonice
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Basso, O.; Lascourreges, J.F.; Le Borgne, F.; Le Goff, C.; Magot, M. Characterization by culture and molecular analysis of the microbial diversity of a deep subsurface gas storage aquifer. Res. Microbiol. 2009, 160, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.E.; Borzenkov, I.A.; Tarasov, A.L.; Milekhina, E.I.; Belyaev, S.S. A microbiological study of an underground gas storage in the process of gas extraction. Microbiology 2007, 76, 461–468. [Google Scholar] [CrossRef]
- Buzek, F.; Onderka, V.; Vančura, P.; Wolf, I. Carbon isotope study of methane production in a town gas storage reservoir. Fuel 1994, 73, 747–752. [Google Scholar] [CrossRef]
- Šmigáň, P.; Greksák, M.; Kozánková, J.; Buzek, F.; Onderka, V.; Wolf, I. Methanogenic bacteria as a key factor involved in changes of town gas stored in an underground reservoir. FEMS Microbiol. Lett. 1990, 73, 221–224. [Google Scholar] [CrossRef]
- Molíková, A.; Vítězová, M.; Vítěz, T.; Buriánková, I.; Huber, H.; Dengler, L.; Hanišáková, N.; Onderka, V.; Urbanová, I. Underground gas storage as a promising natural methane bioreactor and reservoir? J. Energy Storage 2022, 47, 103631. [Google Scholar] [CrossRef]
- Rittmann, S.; Seifert, A.; Herwig, C. Essential prerequisites for successful bioprocess development of biological CH4 production from CO2 and H2. Crit. Rev. Biotechnol. 2015, 35, 141–151. [Google Scholar] [CrossRef]
- Koytsoumpa, E.I.; Bergins, C.; Kakaras, E. The CO2 economy: Review of CO2 capture and reuse technologies. J. Supercrit. Fluids 2018, 132, 3–16. [Google Scholar] [CrossRef]
- Nazir, H.; Louis, C.; Jose, S.; Prakash, J.; Muthuswamy, N.; Buan, M.E.M.; Flox, C.; Chavan, S.; Shi, X.; Kauranen, P.; et al. Is the H2 economy realizable in the foreseeable future ? Part I: H2 production methods. Int. J. Hydrogen Energy 2020, 45, 13777–13788. [Google Scholar] [CrossRef]
- Sampath, P.; Brijesh; Reddy, K.R.; Reddy, C.V.; Shetti, N.P.; Kulkarni, R.V.; Raghu, A.V. Biohydrogen Production from Organic Waste—A Review. Chem. Eng. Technol. 2020, 43, 1240–1248. [Google Scholar] [CrossRef]
- Aguilera, R.F.; Aguilera, R. Revisiting the role of natural gas as a transition fuel. Miner. Econ. 2020, 33, 73–80. [Google Scholar] [CrossRef]
- Pedersen, K.; Arlinger, J.; Ekendahl, S.; Hallbeck, L. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Äspö hard rock laboratory, Sweden. FEMS Microbiol. Ecol. 1996, 19, 249–262. [Google Scholar] [CrossRef]
- Kotelnikova, S.; Pedersen, K. Evidence for methanogenic Archaea and homoacetogenic Bacteria in deep granitic rock aquifers. FEMS Microbiol. Rev. 1997, 20, 339–349. [Google Scholar] [CrossRef]
- Fry, N.K.; Fredrickson, J.K.; Fishbain, S.; Wagner, M.; Stahl, D.A. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl. Environ. Microbiol. 1997, 63, 1498–1504. Available online: https://0-aem-asm-org.brum.beds.ac.uk/content/63/4/1498.short (accessed on 21 May 2019). [CrossRef] [Green Version]
- Shimizu, S.; Akiyama, M.; Ishijima, Y.; Hama, K.; Kunimaru, T.; Naganuma, T. Molecular characterization of microbial communities in fault-bordered aquifers in the Miocene formation of northernmost Japan. Geobiology 2006, 4, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Kimura, H.; Nashimoto, H.; Shimizu, M.; Hattori, S.; Yamada, K.; Koba, K.; Yoshida, N.; Kato, K. Microbial methane production in deep aquifer associated with the accretionary prism in Southwest Japan. ISME J. 2010, 4, 531–541. [Google Scholar] [CrossRef]
- Flynn, T.M.; Sanford, R.A.; Ryu, H.; Bethke, C.M.; Levine, A.D.; Ashbolt, N.J.; Santo Domingo, J.W. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 2013, 13, 146. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Holmfeldt, K.; Hubalek, V.; Lundin, D.; Åström, M.; Bertilsson, S.; Dopson, M. Microbial metagenomes from three aquifers in the Fennoscandian shield terrestrial deep biosphere reveal metabolic partitioning among populations. ISME J. 2016, 10, 1192–1203. [Google Scholar] [CrossRef] [Green Version]
- Frank, Y.A.; Kadnikov, V.V.; Gavrilov, S.N.; Banks, D.; Gerasimchuk, A.L.; Podosokorskaya, O.A.; Merkel, A.Y.; Chernyh, N.A.; Mardanov, A.V.; Ravin, N.V.; et al. Stable and variable parts of microbial community in Siberian deep subsurface thermal aquifer system revealed in a long-term monitoring study. Front. Microbiol. 2016, 7, 2101. [Google Scholar] [CrossRef]
- Kadnikov, V.V.; Frank, Y.A.; Mardanov, A.V.; Beletsky, A.V.; Ivasenko, D.A.; Pimenov, N.V.; Karnachuk, O.V.; Ravin, N.V. Variability of the composition of the microbial community of the deep subsurface thermal aquifer in Western Siberia. Microbiology 2017, 86, 765–772. [Google Scholar] [CrossRef]
- Vigneron, A.; Bishop, A.; Alsop, E.B.; Hull, K.; Rhodes, I.; Hendricks, R.; Head, I.M.; Tsesmetzis, N. Microbial and isotopic evidence for methane cycling in hydrocarbon-containing groundwater from the Pennsylvania region. Front. Microbiol. 2017, 8, 593. [Google Scholar] [CrossRef] [Green Version]
- Evans, P.N.; Boyd, J.A.; Leu, A.O.; Woodcroft, B.J.; Parks, D.H.; Hugenholtz, P.; Tyson, G.W. An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 2019, 17, 219–232. [Google Scholar] [CrossRef]
- Stevens, T.O.; McKinley, J.P.; Fredrickson, J.K. Bacteria Associated with Deep, Alkaline, Anaerobic Groundwaters in Southeast Washington T. Microb. Ecol. 1993, 25, 35–50. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Hedderich, R.; Whitman, W.B. Physiology and biochemistry of the methane-producing Archaea. Prokaryotes 2006, 2, 1050–1079. Available online: http://www.academia.edu/download/63378241/Dworkin_The_Prokaryotes-A_Handbook_on_the_Biology_of_Bacteria_3rd_ed_Vol_220200520-24653-hqh6kx.pdf#page=1100 (accessed on 18 January 2021).
- Lang, K.; Schuldes, J.; Klingl, A.; Poehlein, A.; Daniel, R.; Brune, A. New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of “Candidatus Methanoplasma termitum”. Appl. Environ. Microbiol. 2015, 81, 1338–1352. [Google Scholar] [CrossRef] [Green Version]
- Berghuis, B.A.; Yu, F.B.; Schulz, F.; Blainey, P.C.; Woyke, T.; Quake, S.R. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc. Natl. Acad. Sci. USA 2019, 116, 5037–5044. [Google Scholar] [CrossRef] [Green Version]
- Hattori, S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ. 2008, 23, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Mosbæk, F.; Kjeldal, H.; Mulat, D.G.; Albertsen, M.; Ward, A.J.; Feilberg, A.; Nielsen, J.L. Identification of syntrophic acetate-oxidizing bacteria in anaerobic digesters by combined protein-based stable isotope probing and metagenomics. ISME J. 2016, 10, 2405–2418. [Google Scholar] [CrossRef] [Green Version]
- Hungate, R.E. Chapter IV A Roll Tube Method for Cultivation of Strict Anaerobes. Methods Microbiol. 1969, 3, 117–132. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. A Serum Bottle Modification of the Hungate Technique for Cultivating Obligate Anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [CrossRef]
- Denman, S.E.; Tomkins, N.W.; McSweeney, C.S. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 2007, 62, 313–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinberg, L.M.; Regan, J.M. mcrA-targeted real-time quantitative PCR method to examine methanogen communities. Appl. Environ. Microbiol. 2009, 75, 4435–4442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichler, M.; Coskun, Ö.K.; Ortega-Arbulú, A.S.; Conci, N.; Wörheide, G.; Vargas, S.; Orsi, W.D. A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. Microbiologyopen 2018, 7, e00611. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research 2016, 5, 1496. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Glo, F.O.; Yarza, P. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2016, 2, 16242. [Google Scholar] [CrossRef]
- Whiticar, M.J. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 1999, 161, 291–314. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Baker, B.J.; Saw, J.H.; Lind, A.E.; Lazar, C.S.; Hinrichs, K.U.; Teske, A.P.; Ettema, T.J.G. Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat. Microbiol. 2016, 1, 16002. [Google Scholar] [CrossRef] [Green Version]
- Staniszewska, A.; Kunicka-Styczyńska, A.; Otlewska, A.; Gawor, J.; Gromadka, R.; Żuchniewicz, K.; Ziemiński, K. High-throughput sequencing approach in analysis of microbial communities colonizing natural gas pipelines. Microbiologyopen 2019, 8, e00806. [Google Scholar] [CrossRef]
- Sieber, J.R.; McInerney, M.J.; Gunsalus, R.P. Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation. Annu. Rev. Microbiol. 2012, 66, 429–452. [Google Scholar] [CrossRef]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef]
- Amend, J.P.; Shock, E.L. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 2001, 25, 175–243. [Google Scholar] [CrossRef]
- Yang, G.C.; Zhou, L.; Mbadinga, S.M.; Liu, J.F.; Yang, S.Z.; Gu, J.D.; Mu, B.Z. Formate-dependent microbial conversion of CO2 and the dominant pathways of methanogenesis in production water of high-temperature oil reservoirs amended with bicarbonate. Front. Microbiol. 2016, 7, 365. [Google Scholar] [CrossRef] [Green Version]
- Vítězová, M.; Urbanová, I.; Molíková, A.; Buriánková, I.; Hanišáková, N.; Onderka, V.; Vítěz, T.; Javůrek, J.; Machálková, M. In situ field experiment shows the potential of methanogenic archaea for biomethane production from underground gas storage. Unpublished work.
- Godsy, E.M. Isolation of Methanobacterium bryantii from a deep aquifer by using a novel broth-antibiotic disk method. Appl. Environ. Microbiol. 1980, 39, 1074–1075. [Google Scholar] [CrossRef] [Green Version]
- Kotelnikova, S.; Macario, A.J.L.; Pedersen, K. Methanobacterium subterraneum sp. nov., a new alkaliphilic, eurythermic and halotolerant methanogen isolated from deep granitic groundwater. Int. J. Syst. Bacteriol. 1998, 48, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Qiu, T.L.; Li, X.; Wang, W.D.; Deng, Y.; Yin, X.B.; Zhang, H. Isolation and characterization of Methanoculleus receptaculi sp. nov. from Shengli oil field, China. FEMS Microbiol. Lett. 2008, 285, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Ollivier, B.; Fardeau, M.; Cayol, J.; Magot, M.; Patel, B.K.C.; Prensiep, G.; Garcia, J. Methanocalculus halotolerans. Int. J. Syst. Evol. Microbiol. 1998, 48, 821–828. [Google Scholar]
- Shlimon, A.G.; Friedrich, M.W.; Niemann, H.; Ramsing, N.B.; Finster, K. Methanobacterium aarhusense sp. nov., a novel methanogen isolated from a marine sediment (Aarhus Bay, Denmark). Int. J. Syst. Evol. Microbiol. 2004, 54, 759–763. [Google Scholar] [CrossRef] [Green Version]
- Vanwonterghem, I.; Evans, P.N.; Parks, D.H.; Jensen, P.D.; Woodcroft, B.J.; Hugenholtz, P.; Tyson, G.W. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 2016, 1, 16170. [Google Scholar] [CrossRef] [Green Version]
- Nobu, M.K.; Narihiro, T.; Kuroda, K.; Mei, R.; Liu, W.T. Chasing the elusive Euryarchaeota class WSA2: Genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 2016, 10, 2478–2487. [Google Scholar] [CrossRef]




| Author, Reference | Year | Country | Focus of Study |
|---|---|---|---|
| Šmigán et al. [4] | 1989 | Czech Republic | methanogenic archaea |
| Buzek et al. [4] | 1994 | Czech Republic | microbial methane production |
| Pedersen et al. [11] | 1996 | Sweden | microbial diversity |
| Kotelnikova et al. [12] | 1997 | Sweden | methanogenic archaea, homoacetogenic bacteria |
| Fry et al. [13] | 1997 | USA | microbial diversity |
| Shimizu et al. [14] | 2006 | Japan | microbial diversity |
| Ivanova et al. [2] | 2007 | Russia | microbiological |
| Basso et al. [1] | 2009 | France | microbial diversity |
| Kimura et al. [15] | 2010 | Japan | microbial methane study |
| Flynn et al. [16] | 2013 | USA | functional microbial diversity |
| Wu et al. [17] | 2016 | Sweden | microbial diversity, metabolism |
| Frank et al. [18] | 2016 | Russia | variability in microbial composition |
| Kadnikov et al. [19] | 2017 | Russia | microbial diversity |
| Vigneron et al. [20] | 2017 | USA | microbial methane study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buriánková, I.; Molíková, A.; Vítězová, M.; Onderka, V.; Vítěz, T.; Urbanová, I.; Hanišáková, N.; Černý, M.; Novák, D.; Lochman, J.; et al. Microbial Communities in Underground Gas Reservoirs Offer Promising Biotechnological Potential. Fermentation 2022, 8, 251. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation8060251
Buriánková I, Molíková A, Vítězová M, Onderka V, Vítěz T, Urbanová I, Hanišáková N, Černý M, Novák D, Lochman J, et al. Microbial Communities in Underground Gas Reservoirs Offer Promising Biotechnological Potential. Fermentation. 2022; 8(6):251. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation8060251
Chicago/Turabian StyleBuriánková, Iva, Anna Molíková, Monika Vítězová, Vladimír Onderka, Tomáš Vítěz, Iva Urbanová, Nikola Hanišáková, Martin Černý, David Novák, Jan Lochman, and et al. 2022. "Microbial Communities in Underground Gas Reservoirs Offer Promising Biotechnological Potential" Fermentation 8, no. 6: 251. https://0-doi-org.brum.beds.ac.uk/10.3390/fermentation8060251