Extensive Sampling Provides New Insights into Phylogenetic Relationships between Wild and Domesticated Zanthoxylum Species in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction, PCR Amplification, and Chloroplast DNA Sequencing
2.3. Data Analyses
3. Results
3.1. Sequence Characteristics and Haplotype Detection
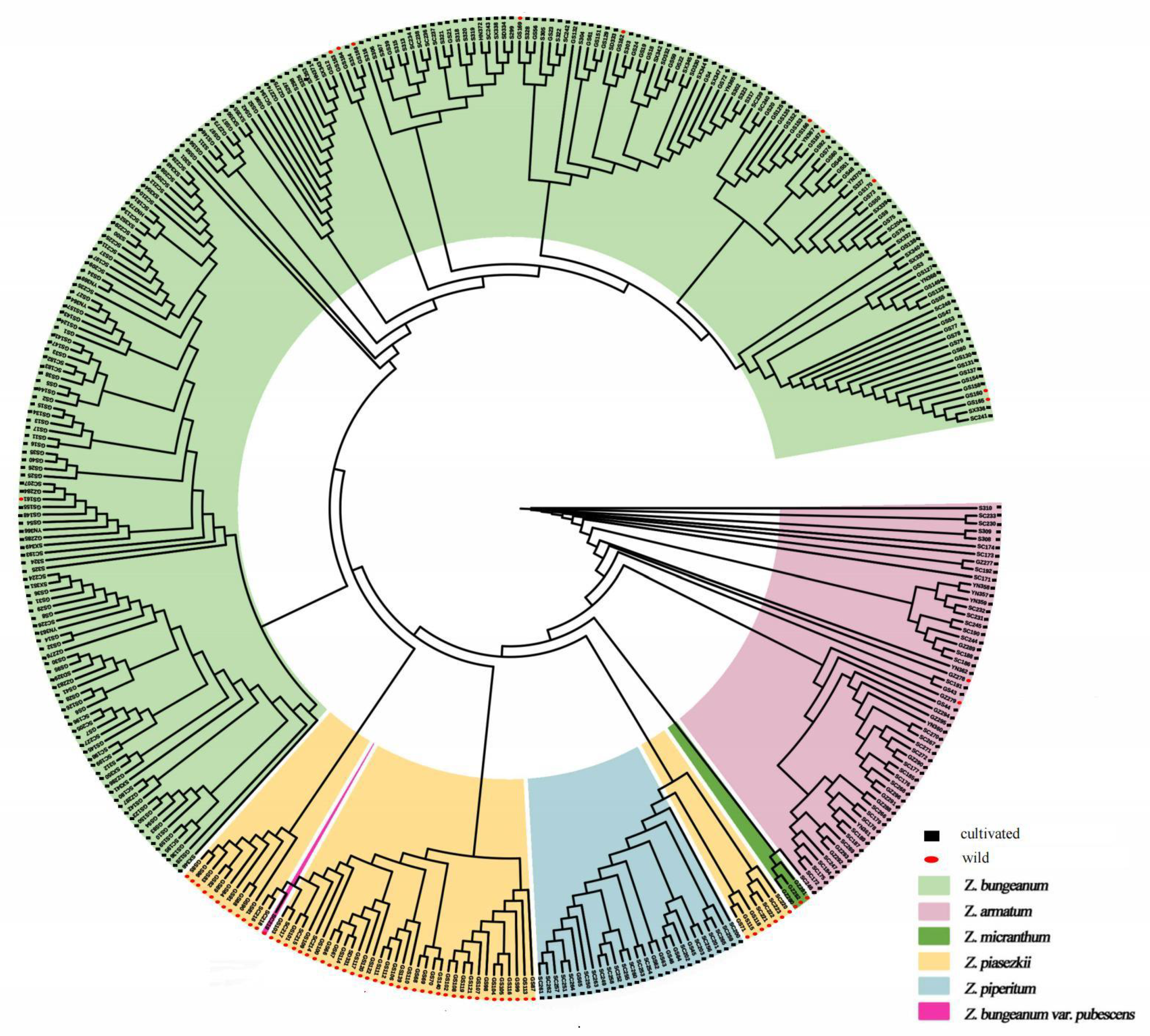
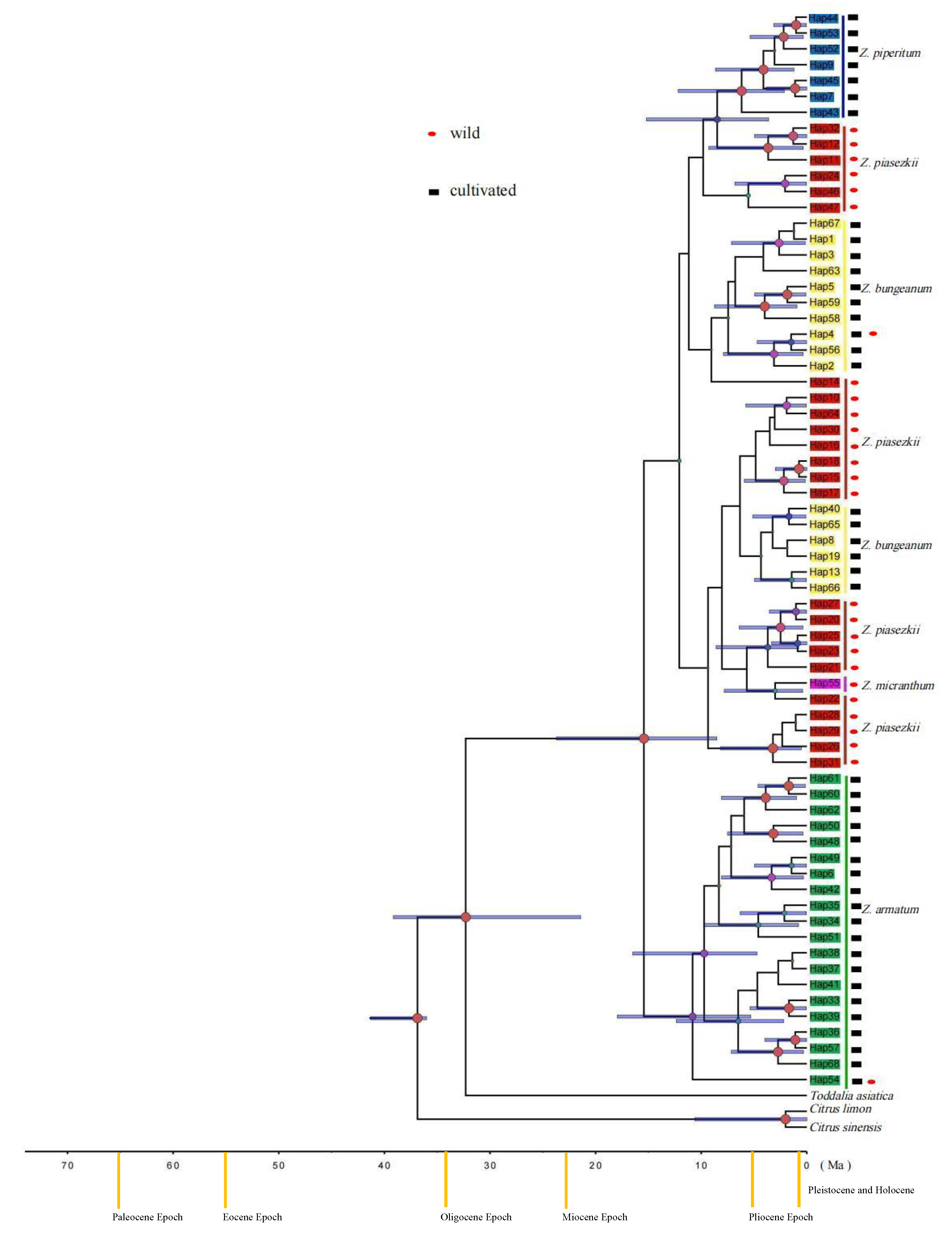
3.2. Genetic Diversity and Phylogenetic Relationships
3.3. Genetic Structure
4. Discussion
4.1. Effect of Human Activity on the Genetic Structure of Zanthoxylum
4.2. The Origin of Cultivated Zanthoxylum Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, W. Research progress of analysis of odor consitituents in Zanthoxylum bungeanum Maxim. China Food Addit. 2021, 32, 192–196. [Google Scholar] [CrossRef]
- Fei, X.T. Analysis of Apomixis Characteristics and Functional Verification of Key Genes in Zanthoxylum bungeanum; Northwest A&F University: Yangling, China, 2021. [Google Scholar]
- Xi, S.Y.; Guo, Y.X.; Ma, X.H.; Zhan, Z.L.; Jin, L. Research progress on chemical constituents and pharmacological effects of Zanthoxylum. West China J. Pharm. Sci. 2021, 36, 717–722. [Google Scholar] [CrossRef]
- Yang, X.F.; Long, Y.Y.; Wu, Y.; Ma, Y.M. Recent progress in research on active componets of Zanthoxylum L. Food Sci. 2017, 39, 303–312. [Google Scholar] [CrossRef]
- Lou, J.R.; Zheng, Z.F.; Li, Y.; Wang, W.X.; Yuan, C. Progress on anti-infectious activities of plants from genus Zanthoxylum. Chin. Tradit. Herb. Drugs 2018, 49, 5477–5484. [Google Scholar] [CrossRef]
- Yuan, H.M.; Peng, C.; Song, Y.; Zhou, C.; Huang, X.; Zhang, Y. Research progress on repellent activities of Zanthoxylum plants. Chin. J. Hyg. Insectic. Equip. 2021, 27, 569–577. [Google Scholar] [CrossRef]
- Pan, S.X.; Pu, B.; Fu, B.N.; Jiang, P.J. Research progress in the sensory classification and detection of numb-taste components of Zanthoxylum. Sci. Technol. Offood Ind. 2017, 38, 347–351. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, Y.F.; Ge, B.G.; Sun, M.; Zhao, Y.; Zhu, W.T. Optimization Extraction Process of Pepper Seed Oil. China Fruit Veg. 2017, 37, 1008–1038. [Google Scholar] [CrossRef]
- Feng, S.; Liu, Z.; Chen, L.; Hou, N.; Yang, T.; Wei, A. Phylogenetic relationships among cultivated Zanthoxylum species in China based on cpDNA markers. Tree Genet. Genomes 2016, 12, 45. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.S.; Wei, X.; Cao, L.R.; Qiao, W.H.; Zhang, W.X.; Yang, Q.W. Origin and evolution of cultivated rice (O. sativa L.) in China based on gene diversity of chloroplast genome. J. Plant Genet. Resour. 2011, 12, 686–693. [Google Scholar] [CrossRef]
- Badenes, M.L.; Parfitt, D.E. Phylogenetic relationships of cultivated Prunus species from an analysis of chloroplast DNA variation. Theor. Appl. Genet. 1995, 90, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.H.; Abe, J.; Kanazawa, A.; Gai, J.Y.; Shimamoto, Y. Identification of sequence variations by PCR-RFLP and its application to the evaluation of cpDNA diversity in wild and cultivated soybeans. Theor. Appl. Genet. 2001, 102, 683–688. [Google Scholar] [CrossRef]
- Wang, C.; Jiao, C.H.; Yang, X.S.; Zhang, W.Y.; Lei, J.; Chai, S.S.; Wang, L.J.; Tian, X.H. Genetic diversity analysis of sweet potato based on chloroplast genes trL-trnF, trnH-psbA and trnT-trnL sequences. J. South Agric. 2021, 52, 1536–1544. [Google Scholar] [CrossRef]
- Van Do, T.; Xu, B.; Gao, X.-F. Molecular phylogeny and character evolution of Flemingia (Leguminosae, Papilionoideae, Phaseoleae, Cajaninae) inferred from three cpDNA and nrITS sequence data. Oesterreichische Bot. Z. 2021, 307, 30. [Google Scholar] [CrossRef]
- Kholina, A.B.; Kozyrenko, M.M.; Artyukova, E.V.; Sandanov, D.V. Modern State of Populations of Endemic Oxytropis Species from Baikal Siberia and Their Phylogenetic Relationships Based on Chloroplast DNA Markers. Russ. J. Genet. 2018, 54, 805–815. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Cai, D.Y.; Daniel, P.; Joseph, P.; Liu, J.; Teng, Y.W. Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol. Phylogenetics Evol. 2014, 80, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets Molecular. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Niu, J.; Liu, Z.; Tian, L.; Wang, X.; Wei, A. Genetic Diversity and Evolutionary Relationships of Chinese Pepper Based on nrDNA Markers. Forests 2020, 11, 543. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, D.J.; Wang, K.; Cong, P.H.; Zhang, C.X.; Li, L.W.; Piao, J.C. Chloroplast DNA variation and genetic evolution of Malus sieversii (Ledeb.) M. Roem. J. Ofplant Genet. Resour. 2020, 21, 579–587. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The Molecular Genetics of Crop Domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.; Zhang, G.R.; Qin, Y.X.; Liu, F.Q.; Tang, S.Q. genetic diversity at chloroplast microsatellites (cpSSR) in Abies fanjingshanensis and A. yuanbaoshanensis. Guihaia 2014, 34, 596–600. [Google Scholar]
- Pusadee, T.; Jamjod, S.; Chiang, Y.; Rerkasem, B.; Schaal, B.A. Genetic structure and isolation by distance in a landrace of Thai rice. Proc. Natl. Acad. Sci. USA 2009, 106, 13880–13885. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Pan, C.H. Study on the distribution and use of Zanthoxylum since Ming and Qing Dynasties-focuses on local Chronicles. China Local Rec. 2019, 84–95+127. [Google Scholar]
- Crandall, K.; Templeton, A. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 1993, 134, 959–969. [Google Scholar] [CrossRef]
- Hang, S.U.N. Tethys Retreat and Himalayas-Hengduanshan mountains uplift and their significance on the origin and development of the Sino-Himalayan elements and Alpine flora. Acta Bot. Yunnanica 2002, 24, 273–288. [Google Scholar] [CrossRef]
- Wang, M.; Shi, Y.; Dong, L.J. Germplasm resources and cultivation research of Zanthoxylum bungeanum. Heilongjiang Sci. 2021, 12, 96–97. [Google Scholar]
- Zhang, Y.; Wang, J.L.; Xia, M.Z.; Zhang, F.Q. Phylogeography of Populus cathayana from the Qinghai-Tibetan Plateau based on cp DNA sequence. J. Biol. 2020, 37, 45–49. [Google Scholar]
- Deng, T.; Wu, F.; Su, T.; Zhou, Z.K. Tibetan Plateau—Evolutionary hub of modern biodiversity formation. Sci. Sin. 2020, 50, 177–193. [Google Scholar]
- Mai, D.H. Development and regional differentiation of the European vegetation during the Tertiary. Plant Syst. Evol. 1989, 162, 79–91. [Google Scholar] [CrossRef]
- Lin, H.R. Preliminary study on the history of Zanthoxylum. J. Sichuan For. Sci. Technol. 1985, 69–73. [Google Scholar] [CrossRef]
- Sun, H.; Li, Z.M. Qinghai-Tibet Plateau uplift and its impact on Tethys flora. Adv. Earth Sci. 2003, 18, 852. [Google Scholar] [CrossRef]



| Province | Cities | Latitude/N | Longitude/E | Elevation/m |
|---|---|---|---|---|
| Gansu | Tianshui | 34.54 | 105.82 | 1399.38 |
| Longnan | 33.46 | 104.96 | 1284.08 | |
| Tibetan Autonomous Prefecture of Gannan | 33.72 | 104.26 | 2291.82 | |
| Hui Autonomous Prefecture of Linxia | 35.73 | 103.12 | 1836.13 | |
| Sichuan | Ya’an | 29.68 | 102.39 | 2025.83 |
| Leshan | 29.56 | 103.76 | 365.01 | |
| Neijiang | 29.58 | 105.06 | 335.00 | |
| Aba Tibetan and Qiang Autonomous Prefecture | 31.76 | 103.75 | 2113.09 | |
| Chengdu | 30.68 | 103.60 | 514.91 | |
| Meishan | 29.93 | 103.44 | 464.66 | |
| Guizhou | Liupanshui | 26.04 | 104.93 | 1446.93 |
| Qianxinan Buyi and Miao Autonomous Prefecture | 25.96 | 105.00 | 1477.01 | |
| Shaanxi | Weinan | 35.44 | 110.21 | 1027.83 |
| Baoji | 33.97 | 106.66 | 1243.09 | |
| Yulin | 39.04 | 111.10 | 934.21 | |
| Shandong | Linyi | 35.71 | 118.09 | 313.53 |
| Zaozhuang | 34.89 | 117.69 | 197.16 | |
| Laiwu | 36.21 | 117.68 | 197.17 | |
| Shanxi | Xinzhou | 39.02 | 113.61 | 1875.00 |
| Yangquan | 38.09 | 113.41 | 948.36 | |
| Yunnan | Kunming | 25.04 | 102.67 | 2112.25 |
| Yuxi | 24.44 | 102.98 | 1643.95 | |
| Henan | Sanmenxia | 34.74 | 111.81 | 476.00 |
| Primer Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Length |
|---|---|---|---|
| matK | MF:CAACACGACTTCCTATACCCAC | MR:CAAATACCAAATCCGCCCTC | 1100 bp |
| ndhH | NF:TATCTGCCTTATGTAACCCGTTG | NR:TTTTGGGGCTTCTACTCTCAC | 816 bp |
| psbB | PF:TATCGTGTTCATACCGTCGT | PR:CGTGTCCAAAAGTAAACCAG | 1351 bp |
| rbcL | RF:CACAAACAGAGACTAAAGCG | RR:CAAAGATCTCGGTCAAAGCA | 1200 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Tian, L.; Tian, J.; Wang, G.; Gong, X.; Feng, S.; Wei, A. Extensive Sampling Provides New Insights into Phylogenetic Relationships between Wild and Domesticated Zanthoxylum Species in China. Horticulturae 2022, 8, 440. https://0-doi-org.brum.beds.ac.uk/10.3390/horticulturae8050440
Chen X, Tian L, Tian J, Wang G, Gong X, Feng S, Wei A. Extensive Sampling Provides New Insights into Phylogenetic Relationships between Wild and Domesticated Zanthoxylum Species in China. Horticulturae. 2022; 8(5):440. https://0-doi-org.brum.beds.ac.uk/10.3390/horticulturae8050440
Chicago/Turabian StyleChen, Xue, Lu Tian, Jieyun Tian, Gang Wang, Xia Gong, Shijing Feng, and Anzhi Wei. 2022. "Extensive Sampling Provides New Insights into Phylogenetic Relationships between Wild and Domesticated Zanthoxylum Species in China" Horticulturae 8, no. 5: 440. https://0-doi-org.brum.beds.ac.uk/10.3390/horticulturae8050440






