Explainable Deep Learning Models in Medical Image Analysis
Abstract
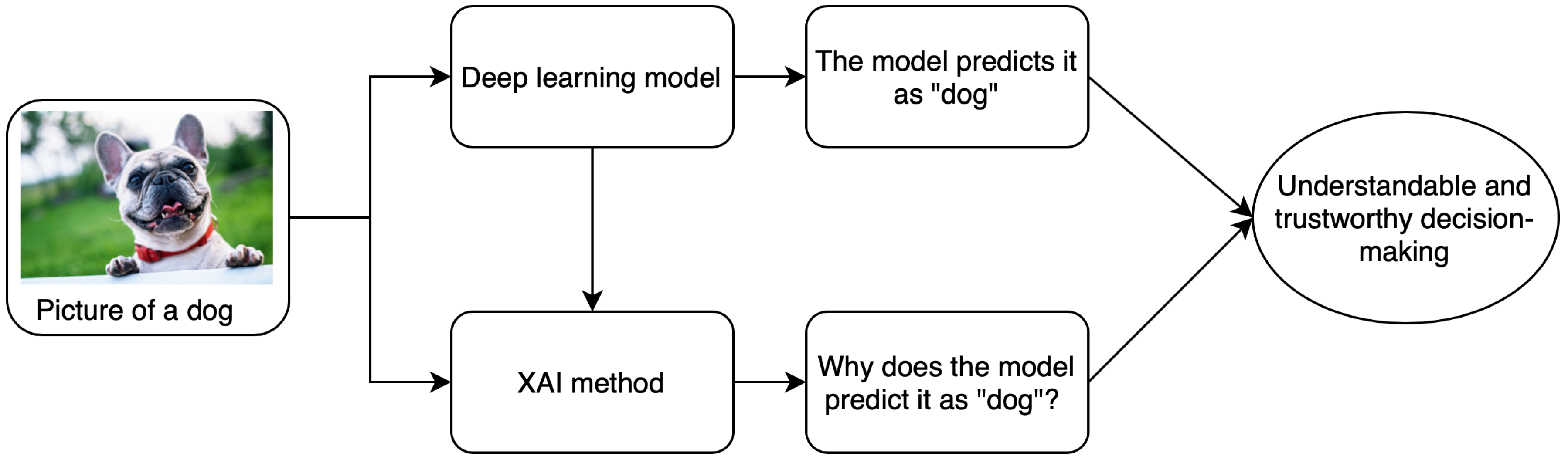
:1. Introduction
2. Taxonomy of Explainability Approaches
2.1. Model Specific vs. Model Agnostic
2.2. Global Methods vs. Local Methods
2.3. Pre-Model vs. In-Model vs. Post-Model
2.4. Surrogate Methods vs. Visualization Methods
3. Explainability Methods—Attribution Based
3.1. Perturbation Based Methods—Occlusion
3.2. Backpropagation Based Methods
4. Applications
4.1. Attribution Based
4.1.1. Brain Imaging
4.1.2. Retinal Imaging
4.1.3. Breast Imaging
4.1.4. CT Imaging
4.1.5. X-ray Imaging
4.1.6. Skin Imaging
4.2. Non-Attribution Based
4.2.1. Attention Based
4.2.2. Concept Vectors
4.2.3. Expert Knowledge
4.2.4. Similar Images
4.2.5. Textual Justification
4.2.6. Intrinsic Explainability
5. Discussion
Funding
Conflicts of Interest
Acronyms
| AI | artificial intelligence |
| AMD | Age-related macular degeneration |
| ASD | autism pectrum disorder |
| CAD | Computer-aided diagnostics |
| CAM | Class activation maps |
| CNN | convolutional neural network |
| CNV | choroidal neovascularization |
| CT | computerized tomography |
| DME | diabetic macular edema |
| DNN | deep neural networks |
| DR | diabetic retinopathy |
| EG | Expressive gradients |
| EHR | electronic healthcare record |
| fMRI | functional magnetic resonance imaging |
| GBP | Guided backpropagation |
| GDPR | General Data Protection Regulation |
| GMM | Gaussian mixture model |
| GradCAM | Gradient weighted class activation mapping |
| GRU | gated recurrent unit |
| HITL | human-in-the-loop |
| IG | Integrated gradients |
| kNN | k nearest neighbors |
| LIFT | Deep Learning Important FeaTures |
| LRP | Layer wise relevance propagation |
| MLP | multi layer perceptron |
| MLS | midline shift |
| MRI | magnetic resonance imaging |
| OCT | optical coherence tomography |
| PCC | Pearson’s correlation coefficient |
| RCV | Regression Concept Vectors |
| ReLU | rectified linear unit |
| RNN | recurrent neural network |
| SHAP | SHapley Additive exPlanations |
| SVM | support vector machines |
| TCAV | Testing Concept Activation Vectors |
| UBS | Uniform unit Ball surface Sampling |
References
- Jo, T.; Nho, K.; Saykin, A.J. Deep learning in Alzheimer’s disease: Diagnostic classification and prognostic prediction using neuroimaging data. Front. Aging Neurosci. 2019, 11, 220. [Google Scholar] [CrossRef] [Green Version]
- Hua, K.L.; Hsu, C.H.; Hidayati, S.C.; Cheng, W.H.; Chen, Y.J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther. 2015, 8, 2015–2022. [Google Scholar]
- Sengupta, S.; Singh, A.; Leopold, H.A.; Gulati, T.; Lakshminarayanan, V. Ophthalmic diagnosis using deep learning with fundus images–A critical review. Artif. Intell. Med. 2020, 102, 101758. [Google Scholar] [CrossRef] [PubMed]
- Leopold, H.; Singh, A.; Sengupta, S.; Zelek, J.; Lakshminarayanan, V. Recent Advances in Deep Learning Applications for Retinal Diagnosis using OCT. In State of the Art in Neural Networks; El-Baz, A.S., Ed.; Elsevier: New York, NY, USA, 2020; in press. [Google Scholar]
- Holzinger, A.; Biemann, C.; Pattichis, C.S.; Kell, D.B. What do we need to build explainable AI systems for the medical domain? arXiv 2017, arXiv:1712.09923. [Google Scholar]
- Stano, M.; Benesova, W.; Martak, L.S. Explainable 3D Convolutional Neural Network Using GMM Encoding. In Proceedings of the Twelfth International Conference on Machine Vision, Amsterdam, The Netherlands, 16–18 November 2019; Volume 11433, p. 114331U. [Google Scholar]
- Moccia, S.; Wirkert, S.J.; Kenngott, H.; Vemuri, A.S.; Apitz, M.; Mayer, B.; De Momi, E.; Mattos, L.S.; Maier-Hein, L. Uncertainty-aware organ classification for surgical data science applications in laparoscopy. IEEE Trans. Biomed. Eng. 2018, 65, 2649–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, T.J.; Ardizzone, L.; Vemuri, A.; Ayala, L.; Gröhl, J.; Kirchner, T.; Wirkert, S.; Kruse, J.; Rother, C.; Köthe, U.; et al. Uncertainty-aware performance assessment of optical imaging modalities with invertible neural networks. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 997–1007. [Google Scholar] [CrossRef] [Green Version]
- Meyes, R.; de Puiseau, C.W.; Posada-Moreno, A.; Meisen, T. Under the Hood of Neural Networks: Characterizing Learned Representations by Functional Neuron Populations and Network Ablations. arXiv 2020, arXiv:2004.01254. [Google Scholar]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef] [Green Version]
- Stiglic, G.; Kocbek, P.; Fijacko, N.; Zitnik, M.; Verbert, K.; Cilar, L. Interpretability of machine learning based prediction models in healthcare. arXiv 2020, arXiv:2002.08596. [Google Scholar]
- Arya, V.; Bellamy, R.K.; Chen, P.Y.; Dhurandhar, A.; Hind, M.; Hoffman, S.C.; Houde, S.; Liao, Q.V.; Luss, R.; Mojsilović, A.; et al. One explanation does not fit all: A toolkit and taxonomy of ai explainability techniques. arXiv 2019, arXiv:1909.03012. [Google Scholar]
- Ying, Z.; Bourgeois, D.; You, J.; Zitnik, M.; Leskovec, J. Gnnexplainer: Generating Explanations for Graph Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems 32, Vancouver, BC, Canada, 8–14 December 2019; Volume 32, pp. 9240–9251. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Maaten, L.V.D.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef] [Green Version]
- Montavon, G.; Lapuschkin, S.; Binder, A.; Samek, W.; Müller, K.R. Explaining nonlinear classification decisions with deep taylor decomposition. Pattern Recognit. 2017, 65, 211–222. [Google Scholar] [CrossRef]
- Ancona, M.; Ceolini, E.; Öztireli, C.; Gross, M. Towards better understanding of gradient-based attribution methods for deep neural networks. arXiv 2017, arXiv:1711.06104. [Google Scholar]
- Alber, M.; Lapuschkin, S.; Seegerer, P.; Hägele, M.; Schütt, K.T.; Montavon, G.; Samek, W.; Müller, K.R.; Dähne, S.; Kindermans, P.J. iNNvestigate neural networks. J. Mach. Learn. Res. 2019, 20, 1–8. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; Springer: Cham, Switzerland; pp. 818–833. [Google Scholar]
- Goodfellow, I.J.; Shlens, J.; Szegedy, C. Explaining and harnessing adversarial examples. arXiv 2014, arXiv:1412.6572. [Google Scholar]
- Lipovetsky, S.; Conklin, M. Analysis of regression in game theory approach. Appl. Stoch. Model. Bus. Ind. 2001, 17, 319–330. [Google Scholar] [CrossRef]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.R.; Samek, W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrikumar, A.; Greenside, P.; Shcherbina, A.; Kundaje, A. Not just a black box: Learning important features through propagating activation differences. arXiv 2016, arXiv:1605.01713. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-Cam: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic Attribution for Deep Networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Voume 70, pp. 3319–3328. [Google Scholar]
- Kindermans, P.J.; Schütt, K.T.; Alber, M.; Müller, K.R.; Erhan, D.; Kim, B.; Dähne, S. Learning how to explain neural networks: Patternnet and patternattribution. arXiv 2017, arXiv:1705.05598. [Google Scholar]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning Important Features through Propagating Activation Differences. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; Voume 70, pp. 3145–3153. [Google Scholar]
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. Smoothgrad: Removing noise by adding noise. arXiv 2017, arXiv:1706.03825. [Google Scholar]
- Chen, H.; Lundberg, S.; Lee, S.I. Explaining Models by Propagating Shapley Values of Local Components. arXiv 2019, arXiv:1911.11888. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How Transferable Are Features in Deep Neural Networks? In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, USA, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Glaucoma diagnosis using transfer learning methods. In Proceedings of the Applications of Machine Learning; International Society for Optics and Photonics (SPIE): Bellingham, WA, USA, 2019; Volume 11139, p. 111390U. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Eitel, F.; Ritter, K.; Alzheimer’s Disease Neuroimaging Initiative (ADNI). Testing the Robustness of Attribution Methods for Convolutional Neural Networks in MRI-Based Alzheimer’s Disease Classification. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support, ML-CDS 2019, IMIMIC 2019; Lecture Notes in Computer Science; Suzuki, K., Ed.; Springer: Cham, Switzerland, 2019; Volume 11797. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Cham, Switzerland, 2018; pp. 106–114. [Google Scholar]
- Sayres, R.; Taly, A.; Rahimy, E.; Blumer, K.; Coz, D.; Hammel, N.; Krause, J.; Narayanaswamy, A.; Rastegar, Z.; Wu, D.; et al. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology 2019, 126, 552–564. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.L.; Kim, J.J.; Kim, J.H.; Kang, Y.K.; Park, D.H.; Park, H.S.; Kim, H.K.; Kim, M.S. Weakly supervised lesion localization for age-related macular degeneration detection using optical coherence tomography images. PLoS ONE 2019, 14, e0215076. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Interpretation of deep learning using attributions: Application to ophthalmic diagnosis. In Proceedings of the Applications of Machine Learning; International Society for Optics and Photonics (SPIE): Bellingham, WA, USA, 2020; in press. [Google Scholar]
- Papanastasopoulos, Z.; Samala, R.K.; Chan, H.P.; Hadjiiski, L.; Paramagul, C.; Helvie, M.A.; Neal, C.H. Explainable AI for medical imaging: Deep-learning CNN ensemble for classification of estrogen receptor status from breast MRI. In Proceedings of the SPIE Medical Imaging 2020: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11314, p. 113140Z. [Google Scholar]
- Lévy, D.; Jain, A. Breast mass classification from mammograms using deep convolutional neural networks. arXiv 2016, arXiv:1612.00542. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet Classification with Deep Convolutional Neural Networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Mordvintsev, A.; Olah, C.; Tyka, M. Inceptionism: Going Deeper into Neural Networks. Google AI Blog. 2015. Available online: https://ai.googleblog.com/2015/06/inceptionism-going-deeper-into-neural.html (accessed on 23 May 2020).
- Couteaux, V.; Nempont, O.; Pizaine, G.; Bloch, I. Towards Interpretability of Segmentation Networks by Analyzing DeepDreams. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2019; pp. 56–63. [Google Scholar]
- Wang, L.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv 2020, arXiv:2003.09871. [Google Scholar]
- Lin, Z.Q.; Shafiee, M.J.; Bochkarev, S.; Jules, M.S.; Wang, X.Y.; Wong, A. Explaining with Impact: A Machine-centric Strategy to Quantify the Performance of Explainability Algorithms. arXiv 2019, arXiv:1910.07387. [Google Scholar]
- Young, K.; Booth, G.; Simpson, B.; Dutton, R.; Shrapnel, S. Deep neural network or dermatologist? In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2019; pp. 48–55. [Google Scholar]
- Van Molle, P.; De Strooper, M.; Verbelen, T.; Vankeirsbilck, B.; Simoens, P.; Dhoedt, B. Visualizing convolutional neural networks to improve decision support for skin lesion classification. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Cham, Switzerland, 2018; pp. 115–123. [Google Scholar]
- Wickstrøm, K.; Kampffmeyer, M.; Jenssen, R. Uncertainty and interpretability in convolutional neural networks for semantic segmentation of colorectal polyps. Med. Image Anal. 2020, 60, 101619. [Google Scholar] [CrossRef] [PubMed]
- Moccia, S.; De Momi, E.; Guarnaschelli, M.; Savazzi, M.; Laborai, A.; Guastini, L.; Peretti, G.; Mattos, L.S. Confident texture-based laryngeal tissue classification for early stage diagnosis support. J. Med. Imaging 2017, 4, 034502. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Nair, B.; Vavilala, M.S.; Horibe, M.; Eisses, M.J.; Adams, T.; Liston, D.E.; Low, D.K.W.; Newman, S.F.; Kim, J.; et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018, 2, 749–760. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Bamba, U.; Pandey, D.; Lakshminarayanan, V. Classification of brain lesions from MRI images using a novel neural network. In Multimodal Biomedical Imaging XV; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11232, p. 112320K. [Google Scholar]
- Zhang, Z.; Xie, Y.; Xing, F.; McGough, M.; Yang, L. Mdnet: A Semantically and Visually Interpretable Medical Image Diagnosis Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6428–6436. [Google Scholar]
- Sun, J.; Darbeha, F.; Zaidi, M.; Wang, B. SAUNet: Shape Attentive U-Net for Interpretable Medical Image Segmentation. arXiv 2020, arXiv:2001.07645. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland; pp. 234–241. [Google Scholar]
- Kim, B.; Wattenberg, M.; Gilmer, J.; Cai, C.; Wexler, J.; Viegas, F.; Sayres, R. Interpretability beyond feature attribution: Quantitative testing with concept activation vectors (tcav). arXiv 2017, arXiv:1711.11279. [Google Scholar]
- Graziani, M.; Andrearczyk, V.; Müller, H. Regression concept vectors for bidirectional explanations in histopathology. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Cham, Switzerland, 2018; pp. 124–132. [Google Scholar]
- Yeche, H.; Harrison, J.; Berthier, T. UBS: A Dimension-Agnostic Metric for Concept Vector Interpretability Applied to Radiomics. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2019; pp. 12–20. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Pisov, M.; Goncharov, M.; Kurochkina, N.; Morozov, S.; Gombolevsky, V.; Chernina, V.; Vladzymyrskyy, A.; Zamyatina, K.; Cheskova, A.; Pronin, I.; et al. Incorporating Task-Specific Structural Knowledge into CNNs for Brain Midline Shift Detection. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2019; pp. 30–38. [Google Scholar]
- Zhu, P.; Ogino, M. Guideline-Based Additive Explanation for Computer-Aided Diagnosis of Lung Nodules. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2019; pp. 39–47. [Google Scholar]
- Codella, N.C.; Lin, C.C.; Halpern, A.; Hind, M.; Feris, R.; Smith, J.R. Collaborative Human-AI (CHAI): Evidence-based interpretable melanoma classification in dermoscopic images. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Cham, Switzerland, 2018; pp. 97–105. [Google Scholar]
- Silva, W.; Fernandes, K.; Cardoso, M.J.; Cardoso, J.S. Towards complementary explanations using deep neural networks. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Cham, Switzerland, 2018; pp. 133–140. [Google Scholar]
- Lee, H.; Kim, S.T.; Ro, Y.M. Generation of Multimodal Justification Using Visual Word Constraint Model for Explainable Computer-Aided Diagnosis. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2019; pp. 21–29. [Google Scholar]
- Biffi, C.; Cerrolaza, J.J.; Tarroni, G.; Bai, W.; De Marvao, A.; Oktay, O.; Ledig, C.; Le Folgoc, L.; Kamnitsas, K.; Doumou, G.; et al. Explainable Anatomical Shape Analysis through Deep Hierarchical Generative Models. IEEE Trans. Med. Imaging 2020. [Google Scholar] [CrossRef] [Green Version]
- Eslami, T.; Raiker, J.S.; Saeed, F. Explainable and Scalable Machine-Learning Algorithms for Detection of Autism Spectrum Disorder using fMRI Data. arXiv 2020, arXiv:2003.01541. [Google Scholar]
- Sha, Y.; Wang, M.D. Interpretable Predictions of Clinical Outcomes with an Attention-Based Recurrent Neural Network. In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MA, USA, 20–23 August 2017; pp. 233–240. [Google Scholar]
- Kaur, H.; Nori, H.; Jenkins, S.; Caruana, R.; Wallach, H.; Wortman Vaughan, J. Interpreting Interpretability: Understanding Data Scientists’ Use of Interpretability Tools for Machine Learning. In Proceedings of the CHI Conference on Human Factors in Computing Systems, Honolulu, HI, USA, 25–30 April 2020; pp. 1–14. [Google Scholar] [CrossRef]
- Arbabshirani, M.R.; Fornwalt, B.K.; Mongelluzzo, G.J.; Suever, J.D.; Geise, B.D.; Patel, A.A.; Moore, G.J. Advanced machine learning in action: Identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit. Med. 2018, 1, 1–7. [Google Scholar] [CrossRef]
- Almazroa, A.; Alodhayb, S.; Osman, E.; Ramadan, E.; Hummadi, M.; Dlaim, M.; Alkatee, M.; Raahemifar, K.; Lakshminarayanan, V. Agreement among ophthalmologists in marking the optic disc and optic cup in fundus images. Int. Ophthalmol. 2017, 37, 701–717. [Google Scholar] [CrossRef]


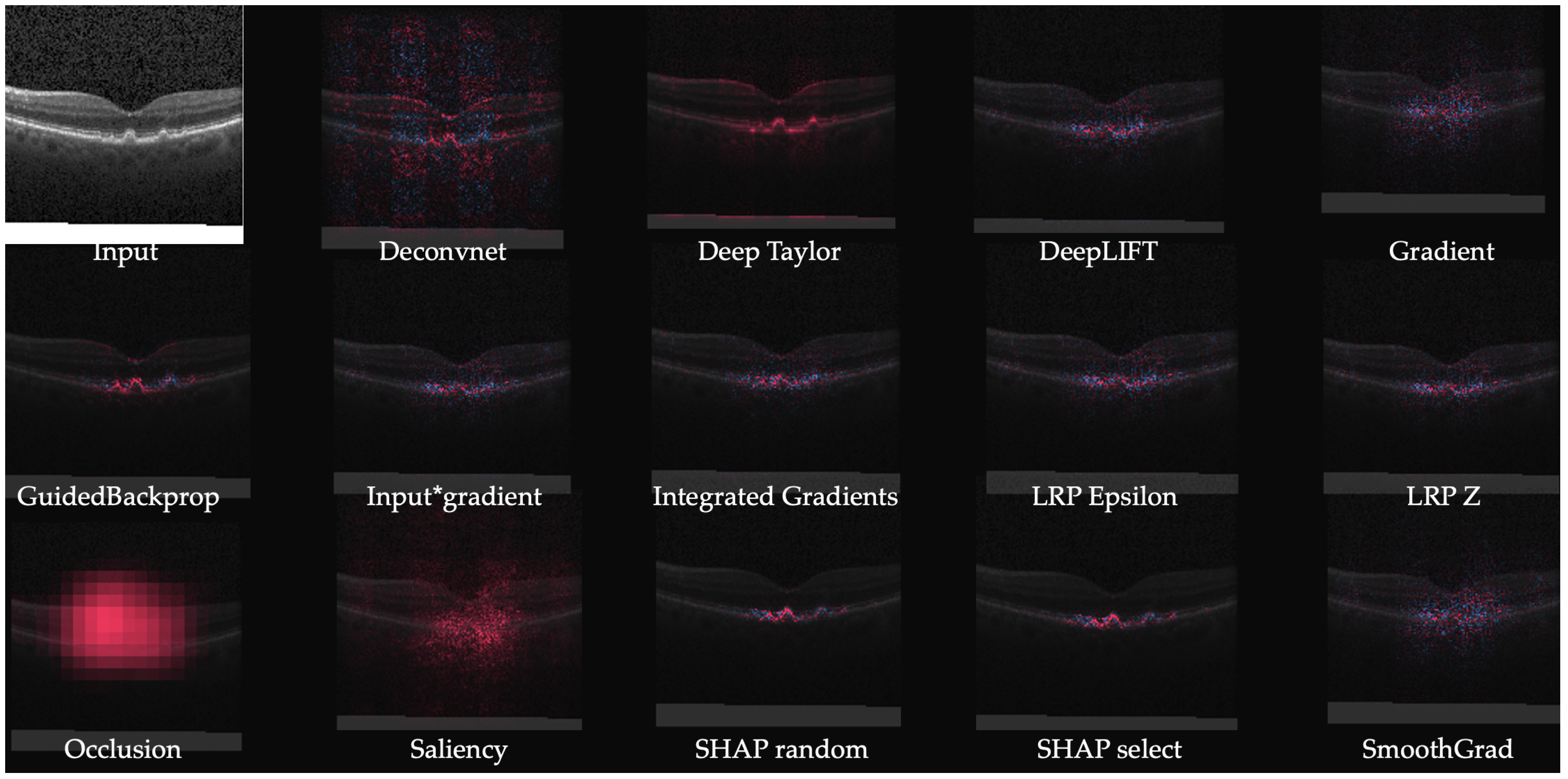

| Method | Description | Notes |
|---|---|---|
| Gradient | Computes the gradient of the output of the target neuron with respect to the input. | The simplest approach but is usually not the most effective. |
| DeConvNet [20] | Applies the ReLU to the gradient computation instead of the gradient of a neuron with ReLU activation. | Used to visualize the features learned by the layers. Limited to CNN models with ReLU activation. |
| Saliency Maps [23] | Takes the absolute value of the partial derivative of the target output neuron with respect to the input features to find the features which affect the output the most with least perturbation. | Can’t distinguish between positive and negative evidence due to absolute values. |
| Guided backpropagation (GBP) [24] | Applies the ReLU to the gradient computation in addition to the gradient of a neuron with ReLU activation. | Like DeConvNet, it is textbflimited to CNN models with ReLU activation. |
| LRP [25] | Redistributes the prediction score layer by layer with a backward pass on the network using a particular rule like the -rule while ensuring numerical stability | There are alternative stability rules and limited to CNN models with ReLU activation when all activations are ReLU. |
| Gradient × input [26] | Initially proposed as a method to improve sharpness of attribution maps and is computed by multiplying the signed partial derivative of the output with the input. | It can approximate occlusion better than other methods in certain cases like multi layer perceptron (MLP) with Tanh on MNIST data [18] while being instant to compute. |
| GradCAM [27] | Produces gradient-weighted class activation maps using the gradients of the target concept as it flows to the final convolutional layer | Applicable to only CNN including those with fully connected layers, structured output (like captions) and reinforcement learning. |
| IG [28] | Computes the average gradient as the input is varied from the baseline (often zero) to the actual input value unlike the Gradient × input which uses a single derivative at the input. | It is highly correlated with the rescale rule of DeepLIFT discussed below which can act as a good and faster approximation. |
| DeepTaylor [17] | Finds a rootpoint near each neuron with a value close to the input but with output as 0 and uses it to recursively estimate the attribution of each neuron using Taylor decomposition | Provides sparser explanations, i.e., focuses on key features but provides no negative evidence due to its assumptions of only positive effect. |
| PatternNet [29] | Estimates the input signal of the output neuron using an objective function. | Proposed to counter the incorrect attributions of other methods on linear systems and generalized to deep networks. |
| Pattern Attribution [29] | Applies Deep Taylor decomposition by searching the rootpoints in the signal direction for each neuron | Proposed along with PatternNet and uses decomposition instead of signal visualization |
| DeepLIFT [30] | Uses a reference input and computes the reference values of all hidden units using a forward pass and then proceeds backward like LRP. It has two variants—Rescale rule and the one introduced later called RevealCancel which treats positive and negative contributions to a neuron separately. | Rescale is strongly related to and equivalent in some cases to -LRP but is not applicable to models involving multiplicative rules. RevealCancel handles such cases and using RevealCancel for convolutional and Rescale for fully connected layers reduces noise. |
| SmoothGrad [31] | An improvement on the gradient method which averages the gradient over multiple inputs with additional noise | Designed to visually sharpen the attributions produced by gradient method using class score function. |
| Deep SHAP [32] | It is a fast approximation algorithm to compute the game theory based SHAP values. It is connected to DeepLIFT and uses multiple background samples instead of one baseline. | Finds attributions for non neural net models like trees, support vector machines (SVM) and ensemble of those with a neural net using various tools in the the SHAP library. |
| Method | Algorithm | Model | Application | Modality |
|---|---|---|---|---|
| Attribution | Gradient*I/P, GBP, LRP, occlusion [36] | 3D CNN | Alzheimer’s detection | Brain MRI |
| GradCAM, GBP [37] | Custom CNN | Grading brain tumor | Brain MRI | |
| IG [38] | Inception-v4 | DR grading | Fundus images | |
| EG [39] | Custom CNN | Lesion segmentation for AMD | Retinal OCT | |
| IG, SmoothGrad [41] | AlexNet | Estrogen receptor status | Breast MRI | |
| Saliency maps [42] | AlexNet | Breast mass classification | Breast MRI | |
| GradCAM, SHAP [49] | Inception | Melanoma detection | Skin images | |
| Activation maps [50] | Custom CNN | Lesion classification | Skin images | |
| DeepDreams [46] | Custom CNN | Segmentation of tumor from liver | CT imaging | |
| GSInquire, GBP, activation maps [47] | COVIDNet CNN | COVID-19 detection | X-ray images | |
| Attention | Mapping between image to reports [56] | CNN & LSTM | Bladder cancer | Tissue images |
| U-Net with shape attention stream [57] | U-net based | Cardiac volume estimation | Cardiac MRI | |
| Concept vectors | TCAV [59] | Inception | DR detection | Fundus images |
| TCAV with RCV [60] | ResNet101 | Breast tumor detection | Breast lymph node images | |
| UBS [61] | SqueezeNet | Breast mass classification | Mammography images | |
| Expert knowledge | Domain constraints [63] | U-net | Brain MLS estimation | Brain MRI |
| Rule-based segmentation, perturbation [64] | VGG16 | Lung nodule segmentation | Lung CT | |
| Similar images | GMM and atlas [6] | 3D CNN | MRI classification | 3D MNIST, Brain MRI |
| Triplet loss, kNN [65] | AlexNet based with shared weights | Melanoma | Dermoscopy images | |
| Monotonic constraints [66] | DNN with two streams | Melanoma detection | Dermoscopy images | |
| Textual justification | LSTM, visual word constraint [67] | Breast mass classification | CNN | Mammography images |
| Intrinsic explainability | Deep Hierarchical Generative Models [68] | Auto-encoders | Classification and segmentation for Alzheimer’s | Brain MRI |
| SVM margin [69] | Hybrid of CNN & SVM | ASD detection | Brain fMRI |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. J. Imaging 2020, 6, 52. https://0-doi-org.brum.beds.ac.uk/10.3390/jimaging6060052
Singh A, Sengupta S, Lakshminarayanan V. Explainable Deep Learning Models in Medical Image Analysis. Journal of Imaging. 2020; 6(6):52. https://0-doi-org.brum.beds.ac.uk/10.3390/jimaging6060052
Chicago/Turabian StyleSingh, Amitojdeep, Sourya Sengupta, and Vasudevan Lakshminarayanan. 2020. "Explainable Deep Learning Models in Medical Image Analysis" Journal of Imaging 6, no. 6: 52. https://0-doi-org.brum.beds.ac.uk/10.3390/jimaging6060052





