Using X-ray Diffraction Techniques for Biomimetic Drug Development, Formulation, and Polymorphic Characterization
Abstract
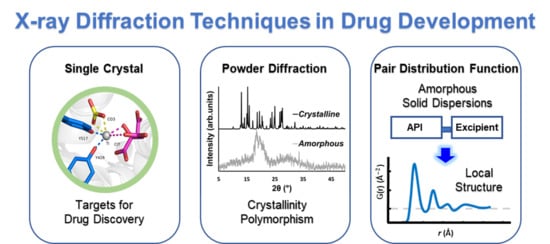
:1. Introduction
2. Significance of X-ray Crystallography in Drug Discovery
2.1. Relevance of the Structural Information of Molecules in Drug Discovery
2.2. Significant Drug Properties Revealed by X-ray Crystallographic Studies
2.3. X-ray Crystallographic Structural Insights Elucidate Promising Biomimetic Drug Candidates
3. Powder X-ray Diffraction (PXRD) in Drug Development, Formulation, and Polymorphic Characterization
4. The Pair Distribution Function (PDF)
4.1. The PDF in Drug Polymorphism
4.2. Pair Distribution Function (PDF) in Amorphous Solid Dispersions (ASD) Formulations
5. Alternative Approaches/Techniques Available for Structure Determination in Drug Discovery
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dabrowiak, J.C. Metals in Medicine, 2nd ed.; Wiley: Hoboken, NJ, USA, 2017; p. 452. [Google Scholar]
- Cleary, E.G.; Beierlein, J.M.; Khanuja, N.S.; McNamee, L.M.; Ledley, F.D. Contribution of NIH funding to new drug approvals 2010–2016. Proc. Natl. Acad. Sci. USA 2018, 115, 2329–2334. [Google Scholar] [CrossRef] [Green Version]
- Bernal, J.D.; Crowfoot, D. X-ray photographs of crystalline pepsin. Nature 1934, 133, 794–795. [Google Scholar] [CrossRef]
- Clark, G.L.; Corrigan, K.E. The crystal structure of insulin. Phys. Rev. 1932, 40, 639. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Dickerson, R.E.; Strandberg, B.E.; Hart, R.G.; Davies, D.R.; Phillips, D.C.; Shore, V.C. Structure of myoglobin: A three-dimensional fourier synthesis at 2 Å. resolution. Nature 1960, 185, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, D.C. The X-ray analysis of the structure of penicillin. Adv. Sci. 1949, 6, 85–89. [Google Scholar] [PubMed]
- Bernal, J.D.; Crowfoot, D.; Fankuchen, I. X-ray crystallography and the chemistry of the steroids. Part I. Philos. Trans. R. Soc. Lond. 1940, 239, 135–182. [Google Scholar] [CrossRef]
- Groom, C.R.; Allen, F.H. The Cambridge Structural Database in retrospect and prospect. Angew. Chem. Int. Ed. Engl. 2014, 53, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekroos, M.; Sjögren, T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. USA 2006, 103, 13682–13687. [Google Scholar] [CrossRef] [Green Version]
- White, S.W.; Zheng, J.; Zhang, Y.M.; Rock. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef]
- Supuran, C.T. Structure-based drug discovery of carbonic anhydrase inhibitors. J. Enzym. Inhib. Med. Chem. 2012, 27, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Gautam, R.; Akam, E.A.; Astashkin, A.V.; Loughrey, J.J.; Tomat, E. Sirtuin inhibitor sirtinol is an intracellular iron chelator. Chem. Commun. 2015, 51, 5104–5107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akam, E.A.; Gautam, R.; Tomat, E. Metal-binding effects of sirtuin inhibitor sirtinol. Supramol. Chem. 2016, 28, 108–116. [Google Scholar] [CrossRef]
- Umena, Y.; Kawakami, K.; Shen, J.R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.; Messinger, J. Reflections on substrate water and dioxygen formation. Biochim. Biophys. Acta 2013, 1827, 1020–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kärkäs, M.D.; Verho, O.; Johnston, E.V.; Åkermark, B. Artificial photosynthesis: Molecular systems for catalytic water oxidation. Chem. Rev. 2014, 114, 11863–12001. [Google Scholar] [CrossRef]
- Yano, J.; Yachandra, V.K. Mn4Ca cluster in photosynthesis: Where and how water is oxidized to dioxygen. Chem. Rev. 2014, 114, 4175–4205. [Google Scholar] [CrossRef]
- Krewald, V.; Retegan, M.; Cox, N.; Messinger, J.; Lubitz, W.; Debeer, S.; Neese, F.; Pantazis, D.A. Metal oxidation states in biological water splitting. Chem. Sci. 2015, 6, 1676–1695. [Google Scholar] [CrossRef] [Green Version]
- Gautam, R.; Astashkin, A.V.; Chang, T.M.; Shearer, J.; Tomat, E. Interactions of metal-based and ligand-based electronic spins in neutral tripyrrindione π dimers. Inorg. Chem. 2017, 56, 6755–6762. [Google Scholar] [CrossRef]
- Gautam, R.; Loughrey, J.J.; Astashkin, A.V.; Shearer, J.; Tomat, E. Tripyrrindione as a redox-active ligand: Palladium(II) coordination in three redox states. Angew. Chem. Int. Ed. Engl. 2015, 54, 14894–14897. [Google Scholar] [CrossRef]
- Chakraborty, S.; Hosen, I.; Ahmed, M.; Shekhar, H.U. Onco-Multi-OMICS approach: A new frontier in cancer research. BioMed Res. Int. 2018, 2018, 9836256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, B.K.; Meerzaman, D. Editorial: Using cancer ‘omics’ to understand cancer. Front. Oncol. 2020, 10, 1201. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.-H.; Snyder, M.P. Omics profiling in precision oncology. Mol. Cell. Proteom. 2016, 15, 2525–2536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, 987. [Google Scholar] [CrossRef]
- Weinberg, R. The Biology of Cancer; Garland Science: New York, NY, USA, 2013; p. 960. [Google Scholar]
- Corcé, V.; Gouin, S.G.; Renaud, S.; Gaboriau, F.; Deniaud, D. Recent advances in cancer treatment by iron chelators. Bioorganic Med. Chem. Lett. 2016, 26, 251–256. [Google Scholar] [CrossRef] [Green Version]
- Steere, A.N.; Byrne, S.L.; Chasteen, N.D.; Mason, A.B. Kinetics of iron release from transferrin bound to the transferrin receptor at endosomal pH. Biochim. Biophys. Acta 2012, 1820, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Dhungana, S.; Taboy, C.H.; Zak, O.; Larvie, M.; Crumbliss, A.L.; Aisen, P. Redox properties of human transferrin bound to its receptor. Biochemistry 2004, 43, 205–209. [Google Scholar] [CrossRef]
- Schlabach, M.R.; Bates, G.W. The synergistic binding of anions and Fe3+ by transferrin. Implications for the interlocking sites hypothesis. J. Biol. Chem. 1975, 250, 2182–2188. [Google Scholar]
- Noinaj, N.; Easley, N.C.; Oke, M.; Mizuno, N.; Gumbart, J.; Boura, E.; Steere, A.N.; Zak, O.; Aisen, P.; Tajkhorshid, E.; et al. Structural basis for iron piracy by pathogenic Neisseria. Nature 2012, 483, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Kolberg, M.; Strand, K.R.; Graff, P.; Andersson, K.K. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta 2004, 1699, 1–34. [Google Scholar] [CrossRef]
- Shao, J.; Liu, X.; Zhu, L.; Yen, Y. Targeting ribonucleotide reductase for cancer therapy. Expert Opin. Ther. Targets 2013, 17, 1423–1437. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Huang, M. Ribonucleotide reductase metallocofactor: Assembly, maintenance and inhibition. Front. Biol. 2014, 9, 104–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stubbe, J.; Ge, J.; Yee, C.S. The evolution of ribonucleotide reduction revisited. Trends Biochem. Sci. 2001, 26, 93–99. [Google Scholar] [CrossRef]
- Keppler, B.K.; Friesen, C.; Moritz, H.G.; Vongerichten, H.; Vogel, E. Tumor-inhibiting bis(β-Diketonato) metal complexes. Budotitane, cis-diethoxybis(1-phenylbutane-1,3-dionato)titanium(IV). In Bioinorganic Chemistry; Springer: Berlin/Heidelberg, Germany, 1991; Volume 78, pp. 97–127. [Google Scholar]
- Harding, M.M.; Mokdsi, G. Antitumour metallocenes: Structure-activity studies and interactions with biomolecules. Curr. Med. Chem. 2000, 7, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Caruso, F.; Rossi, M. Antitumor titanium compounds. Mini Rev. Med. Chem. 2004, 4, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Meléndez, E. Titanium complexes in cancer treatment. Crit. Rev. Oncol. Hematol. 2002, 42, 309–315. [Google Scholar] [CrossRef]
- Buettner, K.M.; Valentine, A.M. Bioinorganic chemistry of titanium. Chem. Rev. 2012, 112, 1863–1881. [Google Scholar] [CrossRef]
- Loza-Rosas, S.A.; Saxena, M.; Delgado, Y.; Gaur, K.; Pandrala, M.; Tinoco, A.D. A ubiquitous metal, difficult to track: Towards an understanding of the regulation of titanium(iv) in humans. Metallomics 2017, 9, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Toney, J.H.; Marks, T.J. Hydrolysis chemistry of the metallocene dichlorides M(.eta.5-C5H5)2Cl2, M = titanium, vanadium, or zirconium. Aqueous kinetics, equilibria, and mechanistic implications for a new class of antitumor agents. J. Am. Chem. Soc. 1985, 107, 947–953. [Google Scholar] [CrossRef]
- Loza-Rosas, S.A.; Vázquez-Salgado, A.; Rivero, K.I.; Negrón, L.J.; Delgado, Y.; Benjamín, J.A.; Vázquez-Maldonado, A.L.; Parks, T.B.; Munet-Colón, C.; Tinoco, A.D. Expanding the therapeutic potential of the iron chelator deferasirox in the development of aqueous stable Ti(IV) anticancer complexes. Inorg. Chem. 2017, 56, 7788–7802. [Google Scholar] [CrossRef]
- Shavit, M.; Peri, D.; Manna, C.M.; Alexander, J.S.; Tshuva, E.Y. Active cytotoxic reagents based on non-metallocene non-diketonato well-defined C2-symmetrical titanium complexes of tetradentate bis(phenolato) ligands. J. Am. Chem. Soc. 2007, 129, 12098–12099. [Google Scholar] [CrossRef] [PubMed]
- Manna, C.M.; Armony, G.; Tshuva, E.Y. Unexpected influence of stereochemistry on the cytotoxicity of highly efficient TiIV salan complexes: New mechanistic insights. Chem. Eur. J. 2011, 17, 14094–14103. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Braitbard, O.; Hochman, J.; Tshuva, E.Y. Insights into molecular mechanism of action of salan titanium(IV) complex with in vitro and in vivo anticancer activity. J. Inorg. Biochem. 2016, 163, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Meker, S.; Braitbard, O.; Hall, M.D.; Hochman, J.; Tshuva, E.Y. Specific design of titanium(IV) phenolato chelates yields stable and accessible, effective and selective anticancer agents. Chem. Eur. J. 2016, 22, 9986–9995. [Google Scholar] [CrossRef]
- Gaur, K.; Vázquez-Salgado, A.M.; Duran-Camacho, G.; Dominguez-Martinez, I.; Benjamín-Rivera, J.A.; Fernández-Vega, L.; Carmona Sarabia, L.; Cruz García, A.; Pérez-Deliz, F.; Méndez Román, J.A.; et al. Iron and copper intracellular chelation as an anticancer grug strategy. Inorganics 2018, 6, 126. [Google Scholar] [CrossRef] [Green Version]
- U.S. National Library of Medicince: ClinicalTrials.gov. ClinicalTrials.gov is a Database of Privately and Publicly Funded Clinical Studies Conducted around the World. Available online: https://clinicaltrials.gov/ct2/home (accessed on 1 December 2020).
- Tinoco, A.D.; Saxena, M.; Sharma, S.; Noinaj, N.; Delgado, Y.; González, E.P.Q.; Conklin, S.E.; Zambrana, N.; Loza-Rosas, S.A.; Parks, T.B. Unusual synergism of transferrin and citrate in the regulation of Ti(IV) speciation, transport, and toxicity. J. Am. Chem. Soc. 2016, 138, 5659–5665. [Google Scholar] [CrossRef]
- Saxena, M.; Loza-Rosas, S.A.; Gaur, K.; Sharma, S.; Perez Otero, S.C.; Tinoco, A.D. Exploring titanium(IV) chemical proximity to iron(III) to elucidate a function for Ti(IV) in the human body. Coord. Chem. Rev. 2018, 363, 109–125. [Google Scholar] [CrossRef]
- Benjamín, J.A.; Cardona-Rivera, A.E.; Vázquez-Maldonado, Á.L.; Dones-Lassalle, C.Y.; Pabón-Colon, H.L.; Rodríguez-Rivera, H.M.; Rodríguez, I.; Gonzalez Espiet, J.C.; Pazol, J.; Pérez, J.; et al. Exploring serum transferrin regulation of nonferric metal therapeutic function and toxicity. Inorganics 2020, 8, 48. [Google Scholar] [CrossRef]
- Guo, M.; Sun, H.; McArdle, H.J.; Gambling, L.; Sadler, P.J. Ti(IV) uptake and release by human serum transferrin and recognition of Ti(IV) transferrin by cancer cells: Understanding the mechanism of action of the anticancer drug titanocene dichloride. Biochemistry 2000, 39, 10023–10033. [Google Scholar] [CrossRef]
- Heinz, U.; Hegetschweiler, K.; Acklin, P.; Faller, B.; Lattmann, R.; Schnebli, H.P. 4-[3,5-Bis(2-hydroxyphenyl)-1,2,4-triazol-1-yl]-benzoic acid: A novel efficient and selective iron(III) complexing agent. Angew. Chem. Int. Ed. 1999, 38, 2568–2570. [Google Scholar] [CrossRef]
- Steinhauser, S.; Heinz, U.; Bartholomä, M.; Weyhermüller, T.; Nick, H.; Hegetschweiler, K. Complex formation of ICL670 and related ligands with Fe-III and Fe-II. Eur. J. Inorg. Chem. 2004, 2004, 4177–4192. [Google Scholar] [CrossRef]
- Crisponi, G.; Nurchi, V.M.; Crespo-Alonso, M.; Sanna, G.; Zoroddu, M.A.; Alberti, G.; Biesuz, R. A speciation study on the perturbing effects of iron chelators on the homeostasis of essential metal ions. PLoS ONE 2015, 10, e0133050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaur, K.; Pérez Otero, S.C.; Benjamin-Rivera, J.A.; Rodriguez, I.; Akam, E.A.; Vazquez-Salgado, A.M.; Loza-Rosas, S.A.; Hernandez, L.; Delgado, Y.; Alicea Fret, N.; et al. Mechanistic insight into the transmetalation inhibition of cellular iron bioavailability by Ti(IV) cTfm complexes. Submitted.
- Ma, R.; Motekaitis, R.J.; Martell, A.E. Stability of metal ion complexes of N,N′-bis(2-hydroxybenzyl)ethylenediamine-N,N′-diacetic acid. Inorg. Chim. Acta 1994, 224, 151–155. [Google Scholar] [CrossRef]
- Tinoco, A.D.; Incarvito, C.D.; Valentine, A.M. Calorimetric, spectroscopic, and model studies provide insight into the transport of Ti(IV) by human serum transferrin. J. Am. Chem. Soc. 2007, 129, 3444–3454. [Google Scholar] [CrossRef]
- Larsen, S.K.; Jenkins, B.G.; Memon, N.G.; Lauffer, R.B. Structure-affinity relationships in the binding of unsubstituted iron phenolate complexes to human serum albumin. Molecular structure of iron(III) N, N’-bis(2-hydroxybenzyl)ethylenediamine-N,N’-diacetate. Inorg. Chem. 1990, 29, 1147–1152. [Google Scholar] [CrossRef]
- Tinoco, A.D.; Thomas, H.R.; Incarvito, C.D.; Saghatelian, A.; Valentine, A.M. Cytotoxicity of a Ti(IV) compound is independent of serum proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 5016–5021. [Google Scholar] [CrossRef] [Green Version]
- Parks, T.B.; Cruz, Y.M.; Tinoco, A.D. Applying the Fe(III) binding property of a chemical transferrin mimetic to Ti(IV) anticancer drug design. Inorg. Chem. 2014, 53, 1743–1749. [Google Scholar] [CrossRef]
- Burley, J.C.; van de Streek, J.; Stephens, P.W. Ampicillin trihydrate from synchrotron powder diffraction data. Acta Crystallogr. Sect. E Struct. Rep. Online 2006, 62, o797–o799. [Google Scholar] [CrossRef]
- Fernandes, P.; Shankland, K.; Florence, A.J.; Shankland, N.; Johnston, A. Solving molecular crystal structures from X-ray powder diffraction data: The challenges posed by gamma-carbamazepine and chlorothiazide N,N,-dimethylformamide (1/2) solvate. J. Pharm. Sci. 2007, 96, 1192–1202. [Google Scholar] [CrossRef]
- Florence, A.J.; Johnston, A.; Fernandes, P.; Shankland, K.; Stevens, H.N.E.; Osmundsen, S.; Mullen, A.B. Powder study of hydrochlorothiazide form II. Acta Crystallogr. Sect. E Struct. Rep. Online 2005, 61, o2798–o2800. [Google Scholar] [CrossRef]
- Shtukenberg, A.G.; Zhu, Q.; Carter, D.J.; Vogt-Maranto, L.; Hoja, J.; Schneider, E.; Song, H.; Pokroy, B.; Polishchuk, I.; Tkatchenko, A.; et al. Powder diffraction and crystal structure prediction identify four new coumarin polymorphs. Chem. Sci. 2017, 8, 4926–4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Censi, R.; Di Martino, P. Polymorph impact on the bioavailability and stability of poorly soluble drugs. Molecules 2015, 20, 18759–18776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilfiker, R. Polymorphism in the Pharmaceutical Industry; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2006. [Google Scholar]
- Elliott, S.R. Medium-range structural order in covalent amorphous solids. Nature 1991, 354, 445–452. [Google Scholar] [CrossRef]
- Cullity, B.D. Elements of X-ray Diffraction; Addison-Wesley: Reading, MA, USA, 1956. [Google Scholar]
- Hancock, B.C.; Zografi, G. Characteristics and significance of the amorphous state in pharmaceutical systems. J. Pharm. Sci. 1997, 86, 1–12. [Google Scholar] [CrossRef]
- Yu, L. Amorphous pharmaceutical solids preparation, characterization. Adv. Drug Deliv. Rev. 2001, 48, 27–42. [Google Scholar] [CrossRef]
- Newman, J.A.; Schmitt, P.D.; Toth, S.J.; Deng, F.; Zhang, S.; Simpson, G.J. Parts per million powder X-ray diffraction. Anal. Chem. 2015, 87, 10950–10955. [Google Scholar] [CrossRef] [Green Version]
- Nunes, C.; Mahendrasingam, A.; Suryanarayanan, R. Quantification of crystallinity in substantially amorphous materials by synchrotron X-ray powder diffractometry. Pharm. Res. 2005, 22, 1942–1953. [Google Scholar] [CrossRef]
- Wagner, C.N.J. Structure of amorphous alloy films. J. Vac. Sci. Technol. 1969, 6, 650–657. [Google Scholar] [CrossRef]
- Thomae, S.L.J.; Prinz, N.; Hartmann, T.; Teck, M.; Correll, S.; Zobel, M. Pushing data quality for laboratory pair distribution function experiments. Rev. Sci. Instruments 2019, 90, 043905. [Google Scholar] [CrossRef]
- Billinge, S.J.L.; Levin, I. The problem with determining atomic structure at the nanoscale. Science 2007, 316, 561–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egami, T.; Billinge, S. Underneath the Bragg peaks. Mater. Today 2003, 6, 57. [Google Scholar] [CrossRef]
- Czernicki, W.; Baranska, M. Carbamazepine polymorphs: Theoretical and experimental vibrational spectroscopy studies. Vib. Spectrosc. 2013, 65, 12–23. [Google Scholar] [CrossRef]
- Billinge, S.J.L.; Dykhne, T.; Juhas, P.; Bozin, E.; Taylor, R.; Florence, A.J.; Shankland, K. Characterisation of amorphous and nanocrystalline molecular materials by total scattering. CrystEngComm 2010, 12, 1366–1368. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Thompson, J.W.; Billinge, S.J.L. PDFgetX2: A GUI-driven program to obtain the pair distribution function from X-ray powder diffraction data. J. Appl. Crystallogr. 2004, 37, 678. [Google Scholar] [CrossRef] [Green Version]
- Davis, T.D. Fingerprinting Analysis of Non-Crystalline Pharmaceutical Compounds Using High Energy X-Rays and the Total Scattering Pair Distribution Function. Ph.D. Thesis, Columbia University, New York, NY, USA, 2011. [Google Scholar]
- Pandey, M.M.; Jaipal, A.; Charde, S.Y.; Goel, P.; Kumar, L. Dissolution enhancement of felodipine by amorphous nanodispersions using an amphiphilic polymer: Insight into the role of drug–polymer interactions on drug dissolution. Pharm. Dev. Technol. 2016, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gao, P. Amorphous pharmaceutical solids: Characterization, stabilization, and development of marketable formulations of poorly soluble drugs with improved oral absorption. Mol. Pharm. 2008, 5, 903–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Ho, C. Amorphous solid dispersions: Utilization and challenges in drug discovery and development. J. Pharm. Sci. 2015, 104, 3237–3258. [Google Scholar] [CrossRef]
- Baghel, S.; Cathcart, H.; O’Reilly, N.J. Polymeric amorphous solid dispersions: A review of amorphization, crystallization, stabilization, solid-state characterization, and aqueous solubilization of biopharmaceutical classification system class II drugs. J. Pharm. Sci. 2016, 105, 2527–2544. [Google Scholar] [CrossRef] [Green Version]
- Kadajji, V.G.; Betageri, G. Water soluble polymers for pharmaceutical applications. Polymers 2011, 3, 1972–2009. [Google Scholar] [CrossRef] [Green Version]
- Sapkal, S.B.; Adhao, V.S.; Thenge, R.R.; Darakhe, R.A.; Shinde, S.A.; Shrikhande, V.N. Formulation and characterization of solid dispersions of etoricoxib using natural polymers. Turk. J. Pharm. Sci. 2020, 17, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Ogaji, I.J.; Nep, I.E.; Audu-Peter, J.D. Advances in natural polymers as pharmaceutical excipients. Pharm. Anal. Acta 2011, 3. [Google Scholar] [CrossRef] [Green Version]
- Shirwaikar, A.; Prabu, S.L.; Kumar, G.A. Herbal excipients in novel drug delivery systems. Indian J. Pharm. Sci. 2008, 70, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murali Mohan Babu, G.V.; Prasad, C.D.S.; Ramana Murthy, K.V. Evaluation of modified gum karaya as carrier for the dissolution enhancement of poorly water-soluble drug nimodipine. Int. J. Pharm. 2002, 234, 1–17. [Google Scholar] [CrossRef]
- Rowe, R.C.; Sheskey, P.J.; Owen, S.C. Handbook of Pharmaceutical Excipients; American Pharmaceutical Association: Washington, DC, USA; Pharmaceutical Press: London, UK, 2006. [Google Scholar]
- Saha, T.; Masum, Z.U.; Mondal, S.K.; Hossain, M.S.; Jobaer, A.; Shahin, R.I.; Fahad, T. Application of natural polymers as pharmaceutical excipients. Global J. Life Sci. Biol. Res. 2018, 4. [Google Scholar] [CrossRef]
- Van Den Mooter, G. The use of amorphous solid dispersions: A formulation strategy to overcome poor solubility and dissolution rate. Drug Discov. Today Technol. 2012, 9, e79–e85. [Google Scholar] [CrossRef]
- Paudel, A.; Worku, Z.A.; Meeus, J.; Guns, S.; Van den Mooter, G. Manufacturing of solid dispersions of poorly water soluble drugs by spray drying: Formulation and process considerations. Int. J. Pharm. 2013, 453, 253–284. [Google Scholar] [CrossRef]
- Forster, A.; Hempenstall, J.; Rades, T. Characterization of glass solutions of poorly water-soluble drugs produced by melt extrusion with hydrophilic amorphous polymers. J. Pharm. Pharmacol. 2001, 53, 303–315. [Google Scholar] [CrossRef]
- Repka, M.A.; Majumdar, S.; Battu, S.K.; Srirangam, R.; Upadhye, S.B. Applications of hot-melt extrusion for drug delivery. Expert. Opin. Drug Deliv. 2008, 5, 1357–1376. [Google Scholar] [CrossRef]
- Serajuddln, A.T.M. Solid Dispersion of poorly water-soluble drugs: Early promises, subsequent problems, and recent breakthroughs. J. Pharm. Sci. 1999, 88, 1058–1066. [Google Scholar] [CrossRef]
- Lui, R. Water-Insoluble Drug Formulation; CRC Press (Interpharm): Boca Raton, FL, USA, 2008. [Google Scholar]
- Janssens, S.; Van den Mooter, G. Review: Physical chemistry of solid dispersions. J. Pharm. Pharmacol. 2009, 61, 1571–1586. [Google Scholar] [CrossRef]
- Shamblin, S.L.; Taylor, L.S.; Zografi, G. Mixing behavior of colyophilized binary systems. J. Pharm. Sci. 1998, 87, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-U.; Krill, S.L.; Wang, Z.; Telang, C. Miscibility/stability considerations in binary solid dispersion systems composed of functional excipients towards the design of multi-component amorphous systems. J. Pharm. Sci. 2009, 98, 4711–4723. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Hu, Y.; Liu, L.; Su, L.; Li, N.; Yu, J.; Tang, B.; Yang, Z. Physical Stability of amorphous solid dispersions: A physicochemical perspective with thermodynamic, kinetic and environmental aspects. Pharm. Res. 2018, 35, 125. [Google Scholar] [CrossRef] [PubMed]
- Crowley, K.J.; Zografi, G. The effect of low concentrations of molecularly dispersed poly (vinylpyrrolidone) on indomethacin crystallization from the amorphous state. Pharm. Res. 2003, 20, 1417–1422. [Google Scholar] [CrossRef]
- Andronis, V.; Zogra, G. Crystal nucleation and growth of indomethacin polymorphs from the amorphous state. J. Non-Cryst. Solids 2000, 21, 236–248. [Google Scholar] [CrossRef]
- Chen, H.; Pui, Y.; Liu, C.; Chen, Z.; Su, C.-C.; Hageman, M.; Hussain, M.; Haskell, R.; Stefanski, K.; Foster, K.; et al. Moisture-induced amorphous phase separation of amorphous solid dispersions: Molecular mechanism, microstructure, and its impact on dissolution performance. J. Pharm. Sci. 2018, 107, 317–326. [Google Scholar] [CrossRef] [Green Version]
- Luebbert, C.; Klanke, C.; Sadowski, G. Investigating phase separation in amorphous solid dispersions via Raman mapping. Int. J. Pharm. 2018, 535, 245–252. [Google Scholar] [CrossRef]
- Purohit, H.S.; Taylor, L.S. Phase separation kinetics in amorphous solid dispersions upon exposure to water. Mol. Pharm. 2015, 12, 1623–1635. [Google Scholar] [CrossRef]
- Rumondor, A.C.F.; Ivanisevic, I.; Bates, S.; Alonzo, D.E.; Taylor, L.S. Evaluation of drug-polymer miscibility in amorphous solid dispersion systems. Pharm. Res. 2009, 26, 2523–2534. [Google Scholar] [CrossRef]
- Takeuchi, I.; Shimakura, K.; Kuroda, H.; Nakajima, T.; Goto, S.; Makino, K. Estimation of crystallinity of nifedipine–polyvinylpyrrolidone solid dispersion by usage of terahertz time-domain spectroscopy and of X-ray powder diffractometer. J. Pharm. Sci. 2015, 104, 4307–4313. [Google Scholar] [CrossRef] [PubMed]
- Nollenberger, K.; Gryczke, A.; Meier, C.; Dressman, J.; Schmidt, M.; Brühne, S. Pair distribution function X-ray analysis explains dissolution characteristics of felodipine melt extrusion products. J. Pharm. Sci. 2009, 98, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Dykhne, T.; Taylor, R.; Florence, A.; Billinge, S.J.L. Data requirements for the reliable use of atomic pair distribution functions in amorphous pharmaceutical fingerprinting. Pharm. Res. 2011, 28, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Araújo, G.; Benmore, C.J.; Byrn, S. Local structure of ion pair interaction in lapatinib amorphous dispersions characterized by synchrotron X-ray diffraction and pair distribution function analysis. Sci. Rep. 2017, 7, 46367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakman, C.; Pestrin, M.; Zafarana, E.; Cantisani, E.; Di Leo, A. Role of lapatinib in the first-line treatment of patients with metastatic breast cancer. Cancer Manag. Res. 2010, 2, 13–25. [Google Scholar]
- Song, Y.; Yang, X.; Chen, X.; Nie, H.; Byrn, S.; Lubach, J.W. Investigation of drug–excipient interactions in lapatinib amorphous solid dispersions using solid-state NMR spectroscopy. Mol. Pharm. 2015, 12, 857–866. [Google Scholar] [CrossRef]
- Mou, Q.; Benmore, C.J.; Yarger, J.L. X-ray Intermolecular Structure Factor(XISF): Separation of intra- and intermolecular interactions from total X-ray scattering data. J. Appl. Crystallogr. 2015, 48, 950–952. [Google Scholar] [CrossRef]
- Keen, D.A. A comparison of various commonly used correlation functions for describing total scattering. J. Appl. Crystallogr. 2001, 34, 172–177. [Google Scholar] [CrossRef]
- Softley, C.A.; Bostock, M.J.; Popowicz, G.M.; Sattler, M. Paramagnetic NMR in drug discovery. J. Biomol. NMR 2020, 74, 287–309. [Google Scholar] [CrossRef]
- Everett, J.R. Drug discovery and development: The role of NMR. eMagRes 2015, 10, 137–150. [Google Scholar] [CrossRef]
- Sugiki, T.; Furuita, K.; Fujiwara, T.; Kojima, C. Current NMR techniques for structure-based drug discovery. Molecules 2018, 23, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emwas, A.H.; Szczepski, K.; Poulson, B.G.; Chandra, K.; McKay, R.T.; Dhahri, M.; Alahmari, F.; Jaremko, L.; Lachowicz, J.I.; Jaremko, M. NMR as a “gold standard” method in drug design and discovery. Molecules 2020, 25, 4597. [Google Scholar] [CrossRef] [PubMed]
- Pellecchia, M.; Bertini, I.; Cowburn, D.; Dalvit, C.; Giralt, E.; Jahnke, W.; James, T.L.; Homans, S.W.; Kessler, H.; Luchinat, C.; et al. Perspectives on NMR in drug discovery: A technique comes of age. Nat. Rev. Drug Discov. 2008, 7, 738–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud, J.-P.; Chari, A.; Ciferri, C.; Liu, W.-T.; Rémigy, H.-W.; Stark, H.; Wiesmann, C. Cryo-EM in drug discovery: Achievements, limitations and prospects. Nat. Rev. Drug Discov. 2018, 17, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Scapin, G.; Potter, C.S.; Carragher, B. Cryo-EM for small molecules discovery, design, understanding, and application. Cell Chem. Biol. 2018, 25, 1318–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudek, A.Z.; Arodz, T.; Galvez, J. Computational methods in developing quantitative structure-activity relationships (QSAR): A review. Comb. Chem. High Throughput Screen. 2006, 9, 213–228. [Google Scholar] [CrossRef] [PubMed]
- McInnes, C. Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 2007, 11, 494–502. [Google Scholar] [CrossRef]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W., Jr. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [Green Version]
- Romano, J.D.; Tatonetti, N.P. Informatics and computational methods in natural product drug discovery: A review and perspectives. Front. Genet. 2019, 10, 368. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Li, X.; Lin, X. A review on applications of computational methods in drug screening and design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [Green Version]
- Ou-Yang, S.-S.; Lu, J.-Y.; Kong, X.-Q.; Liang, Z.-J.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol. Sin. 2012, 33, 1131–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]













| ASD Name | Manufacturer | API | Polymer | Tg of Polymer (°C) |
|---|---|---|---|---|
| Gris-PEG® | Pedinol Pharmacal INC | Griseofulvine | PEG6000 | ~60 |
| Cesamet® | Valeant Pharmaceuticals | Nabilone | PVP | 130–160 |
| Kaletra® | Abbott | Lopinavir, Ritonavir | PVPVA | 106 |
| Sporanox® | Janssen Pharmaceutica | Itraconazole | HPMC | 172 |
| Intelence® | Tibotec | Etravirine | HPMC | 172 |
| Certican® | Novartis | Everolimus | HPMC | 172 |
| Isoptin® SR-E | Abbott | Verapamil | HPC/HPMC | 143–172 |
| Nivadil® | Fujisawa Pharmaceutical Co., Ltd. | Nivaldipine | HPMC | 172 |
| Prograf® | Fujisawa Pharmaceutical Co., Ltd. | Tcrolimus | HPMC | 172 |
| Rezulin® | Developed by Sankyo, manufactured by Parke-Davis division of Warner-Lambert | Troglitazone | PVP | 130–160 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, I.; Gautam, R.; Tinoco, A.D. Using X-ray Diffraction Techniques for Biomimetic Drug Development, Formulation, and Polymorphic Characterization. Biomimetics 2021, 6, 1. https://0-doi-org.brum.beds.ac.uk/10.3390/biomimetics6010001
Rodríguez I, Gautam R, Tinoco AD. Using X-ray Diffraction Techniques for Biomimetic Drug Development, Formulation, and Polymorphic Characterization. Biomimetics. 2021; 6(1):1. https://0-doi-org.brum.beds.ac.uk/10.3390/biomimetics6010001
Chicago/Turabian StyleRodríguez, Israel, Ritika Gautam, and Arthur D. Tinoco. 2021. "Using X-ray Diffraction Techniques for Biomimetic Drug Development, Formulation, and Polymorphic Characterization" Biomimetics 6, no. 1: 1. https://0-doi-org.brum.beds.ac.uk/10.3390/biomimetics6010001