Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Study Site
2.2. Soil Sampling and Properties
2.3. Chemical Analyses
2.4. DNA Extraction and Sequencing
2.5. Bioinformatic Analysis
3. Results
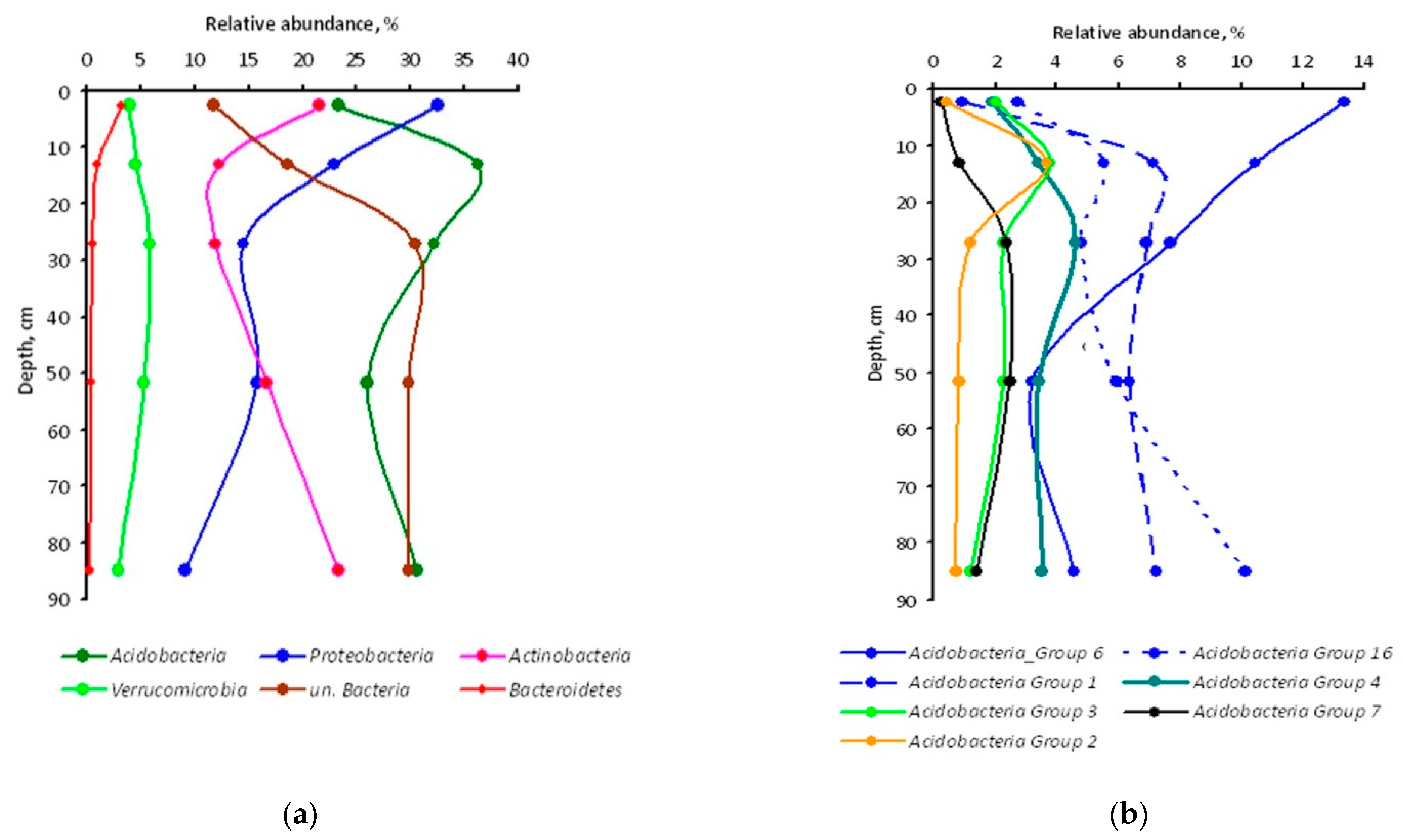
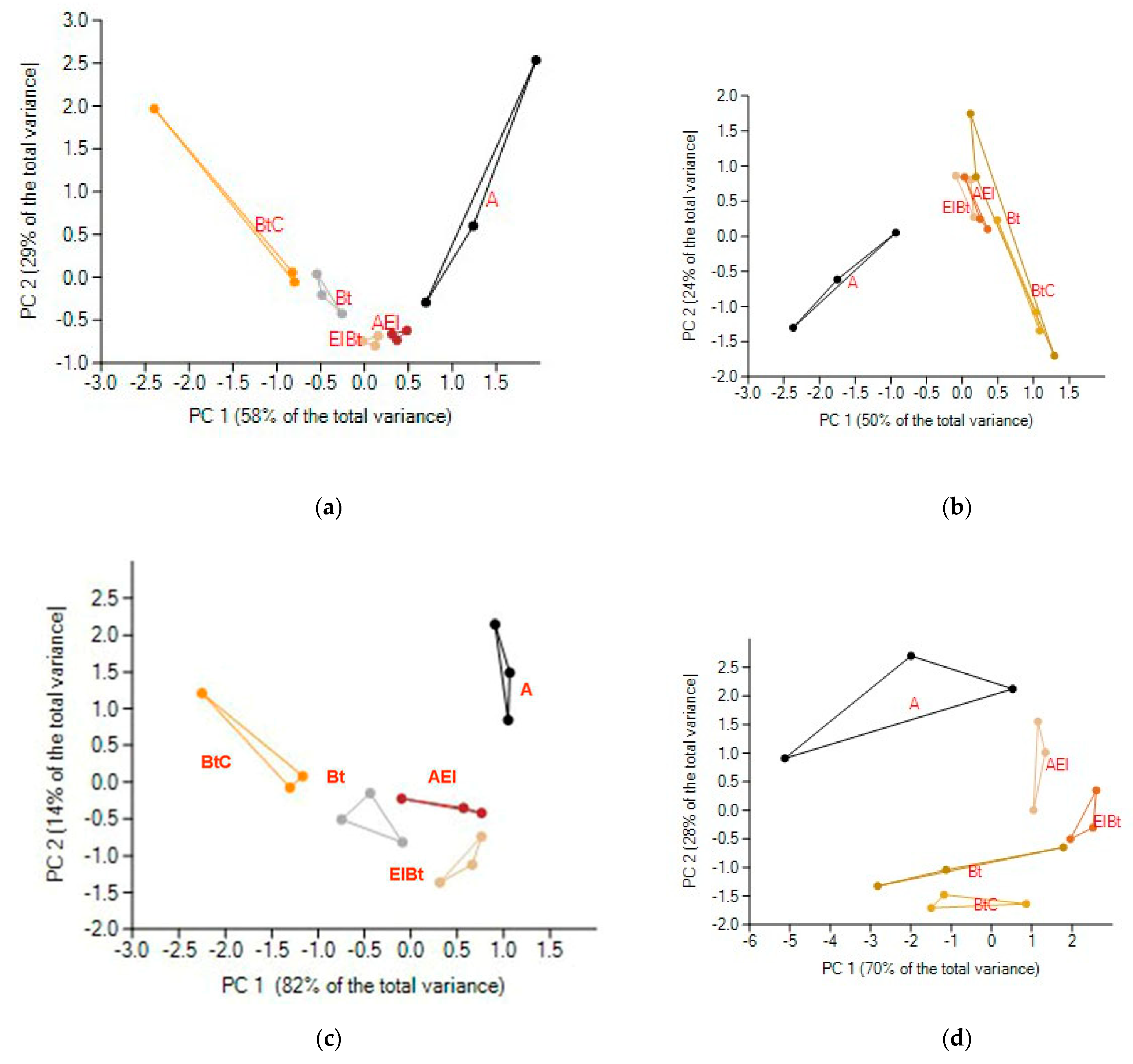
3.1. Bacterial Diversity
3.2. Fungal Diversity
3.3. Bacterial and Fungal α-Diversity Indices
4. Discussion
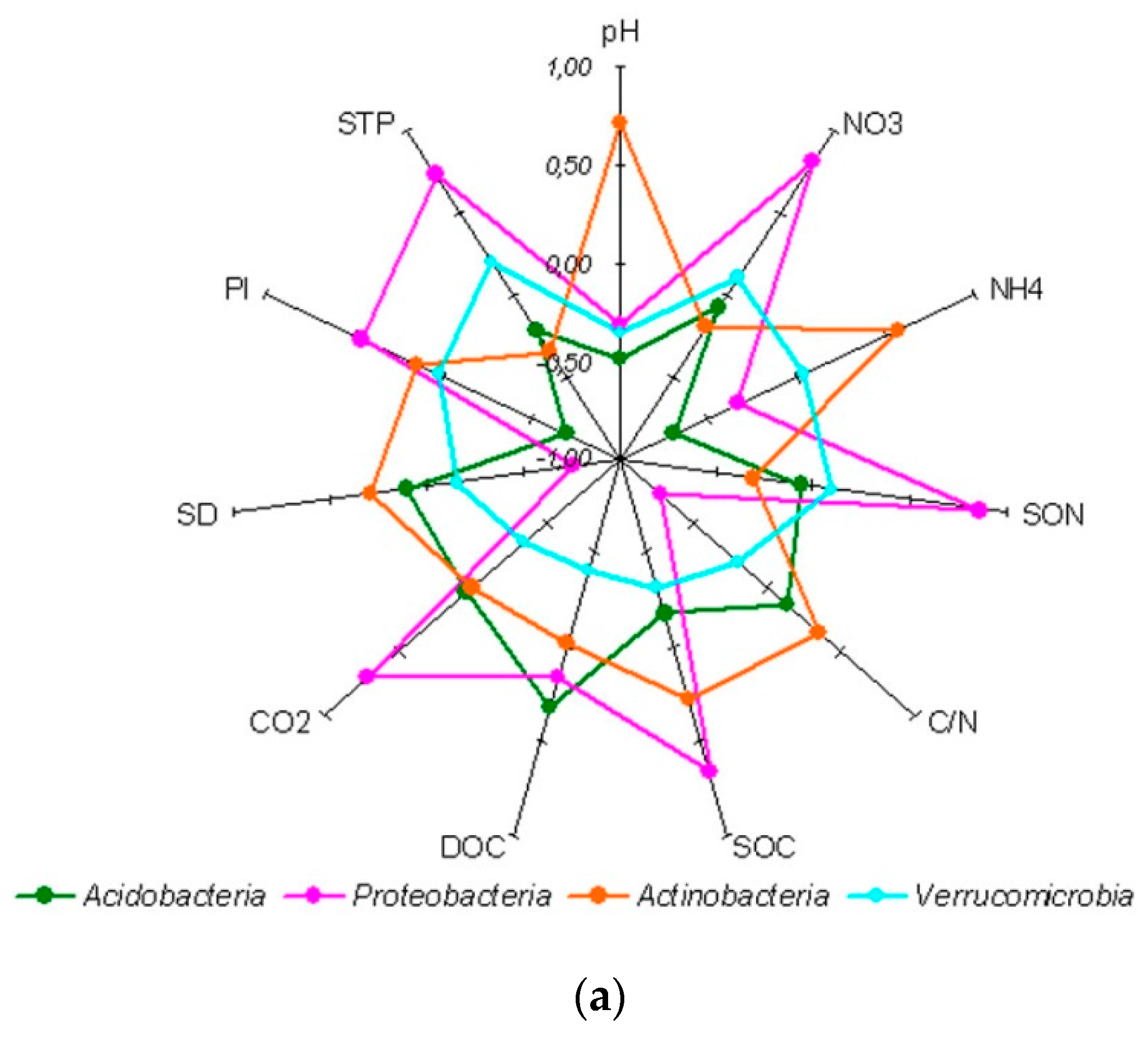
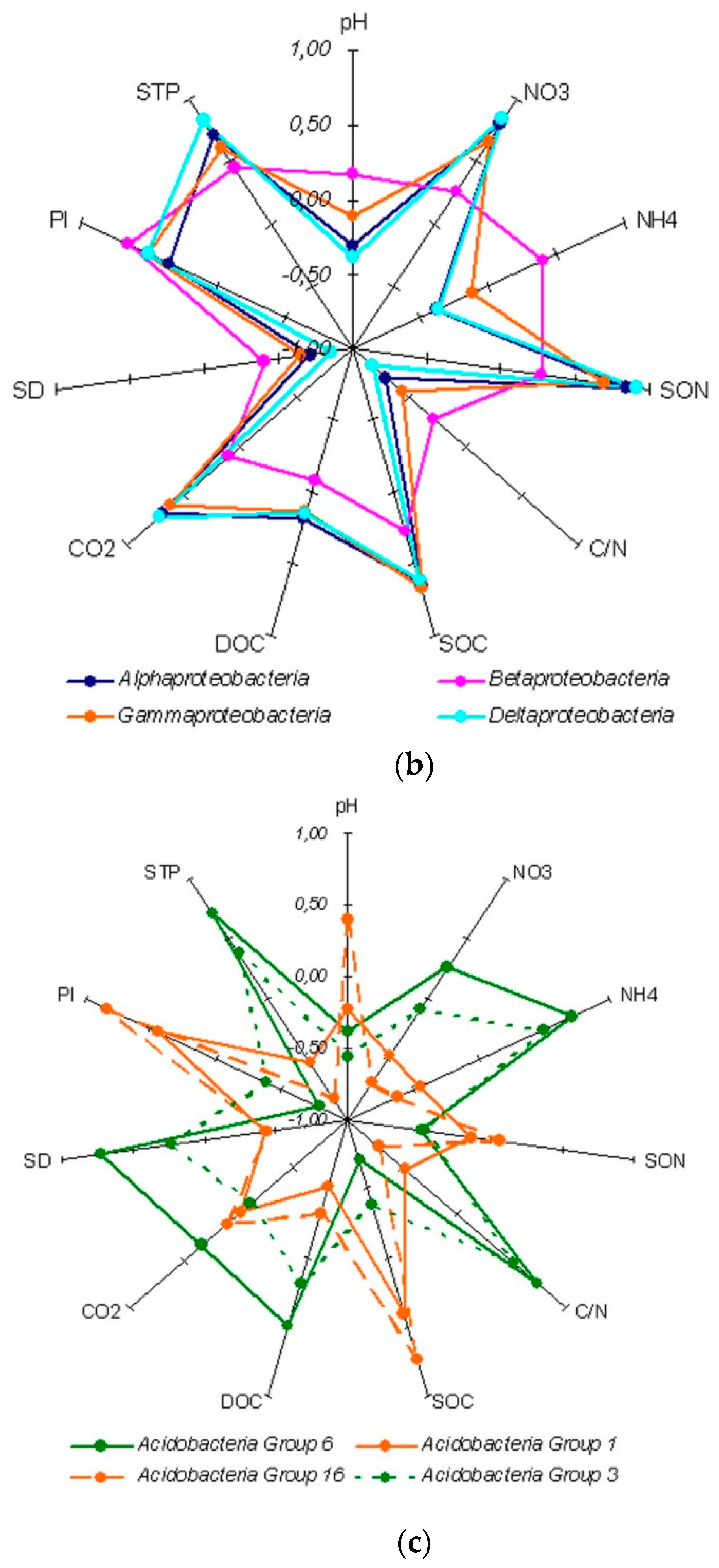
4.1. Bacterial Assemblages in Soil Genetic Horizons
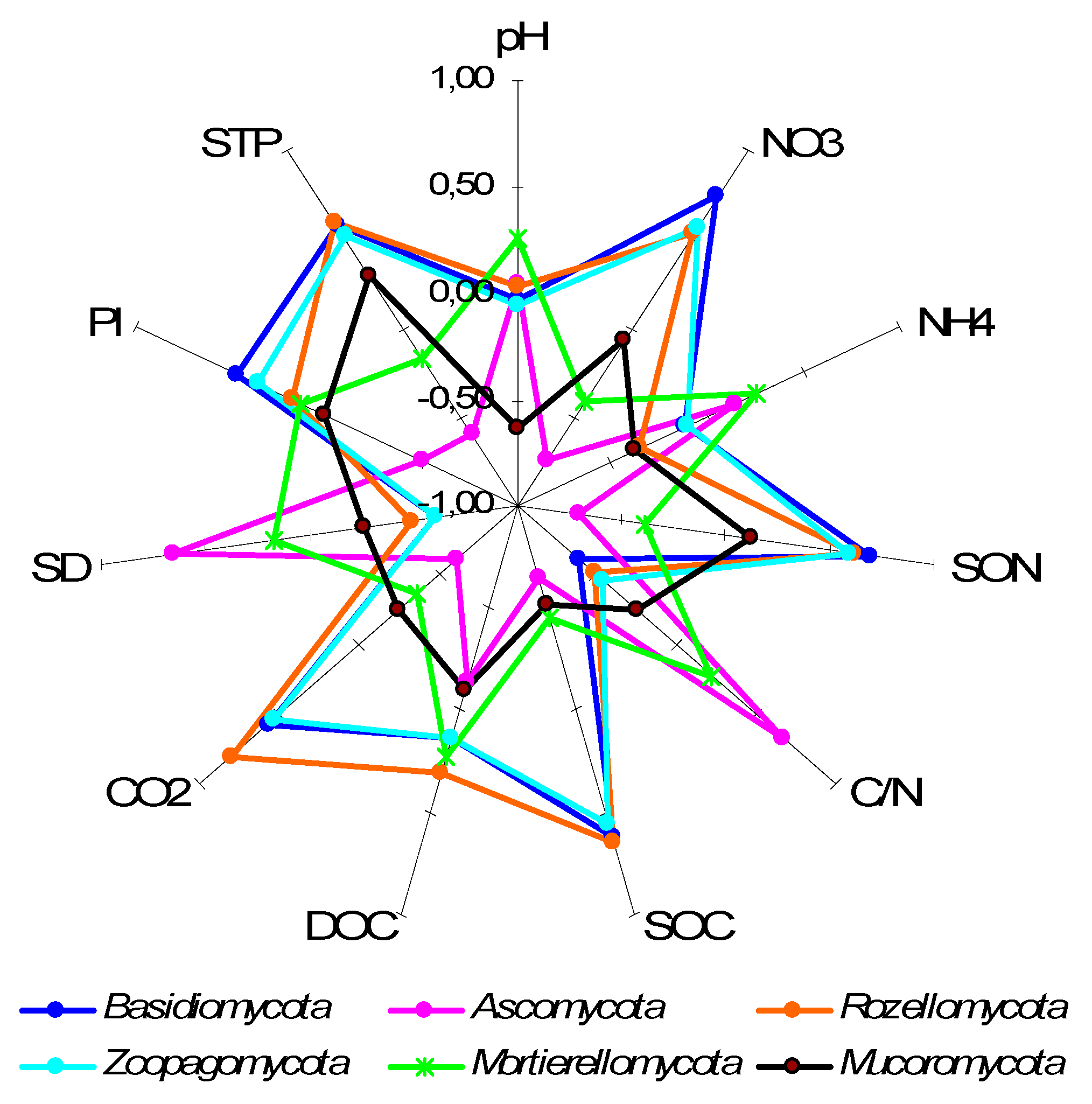
4.2. Fungal Assemblages in Soil Genetic Horizons
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coleman, D.C. Through a ped darkly—An ecological assessment of root soil-microbial-faunal interactions. In Ecological Interactions in the Soil: Plants, Microbes and Animals; Fitter, A.H., Atkinson, D., Read, D.J., Usher, M.B., Eds.; Blackwells: Oxford, UK, 1985; pp. 1–21. [Google Scholar]
- Goldmann, K.; Schröter, K.; Pena, R.; Schöning, I.; Schrumpf, M.; Buscot, F.; Polle, A.; Wubet, T. Divergent habitat filtering of root and soil fungal communities in temperate beech forests. Sci. Rep. 2016, 6, 31439. [Google Scholar] [CrossRef] [PubMed]
- FAO. Global Forest Resources Assessment-2015. Country Report. Russian Federation. 2015. Available online: http://www.fao.org/3/a-az316e.pdf (accessed on 13 December 2019).
- Baldrian, P. Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 2017, 41, 109–130. [Google Scholar] [CrossRef] [Green Version]
- Xue, P.P.; Carrillo, Y.; Pino, V.; Minasny, B.; McBratney, A.B. Soil Properties Drive Microbial Community Structure in a Large Scale Transect in South Eastern Australia. Sci. Rep. 2018, 8, 11725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shishov, L.L.; Tonkonogov, V.D.; Lebedeva, I.I.; Gerasimoiva, M.I. (Eds.) Classification and Diagnostics of Soils in Russia, 1st ed.; Oykumena Pubs: Moscow, Russian, 2004. (In Russian) [Google Scholar]
- IUSS Working Group. WRB, World Reference Base for Soil Resources 2014, Update 2015: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; FAO: Rome, Italy, 2014. [Google Scholar]
- Rabinowitz, H.; Vogel, S. (Eds.) Style and Usage in Earth Science and Environmental Science. In The Manual of Scientific Style; Academic Press/Elsevier: Amsterdam, The Netherlands, 2009; pp. 427–468. [Google Scholar] [CrossRef]
- Carter, M.R.; Gregorich, E.G. (Eds.) Soil Sampling and Methods of Analysis, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Pell, M.; Stenstrom, L.; Granhall, U. Soil respiration. In Microbiological Methods for Assessing Soil Quality; CABI International: Wallingford, UK, 2005; pp. 117–126. [Google Scholar]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igolkina, A.A.; Grekhov, G.A.; Pershina, E.V.; Samosorova, G.G.; Leunova, V.M.; Semenova, A.N.; Baturina, O.A.; Kabilov, M.R.; Andronov, E.E. Identifying components of mixed and contaminated soil samples by detecting specific signatures of control 16S rRNA libraries. Ecol. Ind. 2018, 94, 446–453. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. SINTAX, a Simple Non-Bayesian Taxonomy Classifier for 16S and ITS Sequences. 2016. Available online: http://https://www.biorxiv.org/content/10.1101/074161v1 (accessed on 24 February 2021).
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UNITE Community: UNITE USEARCH/UTAX Release for Fungi Version 18.11.2018. UNITE Community. Available online: https://0-doi-org.brum.beds.ac.uk/10.15156/BIO/786345/ (accessed on 24 February 2021).
- Fauth, E.; Bernardo, J.; Camara, M.; Resetarits, W.J., Jr.; Van Buskirk, J.; McCollum, S.A. Simplifying the Jargon of Community Ecology: A Conceptual Approach. Am. Nat. 1996, 147, 282–286. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Hughes, J.B.; Hellmann, J.J. The Application of Rarefaction Techniques to Molecular Inventories of Microbial Diversity. Methods Enzymol. 2005, 397, 292–308. [Google Scholar] [CrossRef]
- Bai, X.X.; Shi, R.J.; You, Y.M.; Sheng, H.F.; Han, S.Q.; Zhang, Y. Bacterial community structure and diversity in soils of different forest ages and types in Bao- tianman forest, Henan Province, China. Ying Yong Sheng Tai Xue Bao 2015, 26, 2273–2281. (In Chinese) [Google Scholar] [PubMed]
- Wei, H.; Peng, C.; Yang, B.; Song, H.; Li, Q.; Jiang, L.; Wei, G.; Wang, K.; Wang, H.; Liu, S.; et al. Contrasting Soil Bacterial Community, Diversity, and Function in Two Forests in China. Front. Microbiol. 2018, 9, 1693. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, N.T. Contrasting soil bacterial community structure between the phyla Acidobacteria and Proteobacteria in tropical Southeast Asian and temperate Japanese forests. Genes Genet. Syst. 2015, 90, 61–77. [Google Scholar] [CrossRef] [Green Version]
- Stroobants, A.; Degrune, F.; Olivier, C.; Muys, C.; Roisin, C.; Colinet, G.; Bodson, B.; Portetelle, D.; Vandenbol, M. Diversity of Bacterial Communities in a Profile of a Winter Wheat Field: Known and Unknown Members. Microbiol. Ecol. 2014, 68, 822. [Google Scholar] [CrossRef]
- Baldrian, P. The known and the unknown in soil microbial ecology. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, A.P.A.; Andrade, P.A.M.; Bini, D.; Durrer, A.; Robin, A.; Bouillet, J.P.; Andreote, F.D.; Cardoso, E.J.B.N. Shifts in the bacterial community composition along deep soil profiles in monospecific and mixed stands of Eucalyptus grandis and Acacia mangium. PLoS ONE 2017, 12, e0180371. [Google Scholar] [CrossRef] [Green Version]
- Siles, J.A.; Margesin, R. Seasonal soil microbial responses are limited to changes in functionality at two Alpine forest sites differing in altitude and vegetation. Sci. Rep. 2017, 7, 2204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, C.; Xu, C.Y.; Wang, Q.; Zhang, F.; Ma, W.G.; He, W.X.; Hou, L.; Geng, Z.C. Patterns of Bacterial Community Through Soil Depth Profiles and Its Influencing Factors Under Betula albosinensis Burkill in the Xinjiashan Forest Region of Qinling Mountains. Huan Jing Ke Xue. 2017, 38, 3010–3019. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Chen, W.; Wang, J.; Du, N.; Li, Q.; Wei, G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 2018, 6, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichorst, S.A.; Trojan, D.; Roux, S.; Herbold, C.; Rattei, T.; Woebken, D. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 2018, 20, 1041–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Cong, J.; Lu, H.; Li, G.; Qu, Y.; Su, X.; Zhou, J.; Li, D. Community structure and elevational diversity patterns of soil Acidobacteria. J. Environ. Sci. (China) 2014, 26, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Pankratov, T.A.; Serkebaeva, Y.M.; Kulichevskaya, I.S.; Liesack, W.; Dedysh, S.N. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2008, 2, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 2011, 77, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domeignoz-Horta, L.A.; DeAngelis, K.M.; Pold, G. Draft Genome Sequence of Acidobacteria Group 1 Acidipila sp. Strain EB88, Isolated from Forest Soil. Microbiol. Resour. Announ. 2019, 8, e01464-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental factors affect Acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielak, A.M.; van Veen, J.A.; Kowalchuk, G.A. Comparative analysis of acidobacterial genomic fragments from terrestrial and aquatic metagenomic libraries, with emphasis on Acidobacteria subdivision 6. Appl. Environ. Microbiol. 2010, 76, 6769–6777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pold, G.; Billings, A.F.; Blanchard, J.L.; Burkhardt, D.B.; Frey, S.D.; Melillo, J.M.; Schnabel, J.; van Diepen, L.T.; DeAngelis, K.M. Long-Term Warming Alters Carbohydrate Degradation Potential in Temperate Forest Soils. Appl. Environ. Microbiol. 2016, 82, 6518–6530. [Google Scholar] [CrossRef] [Green Version]
- VanInsberghe, D.; Maas, K.R.; Cardenas, E.; Strachan, C.R.; Hallam, S.J.; Mohn, W.W. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J. 2015, 9, 2435–2441. [Google Scholar] [CrossRef] [Green Version]
- Naumova, N.B.; Kuznetsova, G.V.; Alikina, T.Y.; Kabilov, M.R. Bacterial 16S DNA diversity in the rhizosphere soil of two pine species. Biomics 2015, 7, 127–136. [Google Scholar]
- Naumova, N.B.; Alikina, T.Y.; Kuznetsova, G.V. Biodiversity of bacterial assemblages in the Haplic Cambisol under Korean pine. J. Soils Environ. 2018, 1, 151–169. (In Russian) [Google Scholar] [CrossRef]
- Will, C.; Thürmer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M.; Gutknecht, J.; Wubet, T.; Buscot, F.; Daniel, R. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steger, K.; Kim, A.T.; Ganzert, L.; Grossart, H.-P.; Smart, D.R. Floodplain soil and its bacterial composition are strongly affected by depth. FEMS Microbiol. Ecol. 2019, fiz014. [Google Scholar] [CrossRef]
- Du, C.; Geng, Z.; Wang, Q.; Zhang, T.; He, W.; Hou, L.; Wang, Y. Variations in bacterial and fungal communities through soil depth profiles in a Betula albosinensis forest. J. Microbiol. 2017, 55, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Boberg, J.; Cleary, M.; Bruelheide, H.; Hönig, L.; Koricheva, J.; Stenlid, J. Foliar fungi of Betula pendula: Impact of tree species mixtures and assessment methods. Sci. Rep. 2017, 7, 41801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [Green Version]
- Jakucs, E.; Eros-Honti, Z. Morphological-anatomical characterization and identification of Tomentella ectomycorrhizas. Mycorrhiza 2008, 18, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.; Perry, B.A.; Dranginis, A.W.; Pfister, D.H. A phylogeny of the highly diverse cup-fungus family Pyronemataceae, Pezizomycetes, Ascomycota clarifies relationships and evolution of selected life history traits. Mol. Phyl. Evol. 2013, 67, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Hansen, K.; Perry, B.A.; Kjøller, R. Molecular and morphological diversity of pezizalean ectomycorrhiza. New Phytol. 2006, 170, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.-L.; Mortimer, P.E.; Slik, J.W.F.; Zou, X.-M.; Xu, J.; Feng, W.-T.; Qiao, L. Variation in forest soil fungal diversity along a latitudinal gradient. Fungal Div. 2014, 64, 305–315. [Google Scholar] [CrossRef]
- Hartemink, A.E.; Zhang, Y.; Bockheim, J.G.; Curi, N.; Silva, S.H.G.; Grauer-Gray, J.; Lowe, D.J.; Krasilnikov, P. Soil horizon variation: A review. Adv. Agron. 2020, 160, 125–185. [Google Scholar] [CrossRef]




| Property | Soil Genetic Horizon | ||||
|---|---|---|---|---|---|
| A ¥ | AEl | ElBt | Bt | BtC | |
| Depth cm | 0–5 | 5–21 | 21–33 | 33–70 | 70–100 |
| Soil bulk density, g cm−3 soil | 0.42 a # | 1.14 b | 1.13 b | 1.33 c | 1.47 d |
| pH | 6.5 b | 6.4 a | 6.3 a | 6.5 b | 6.6 b |
| DOC, µg kg-1 soil | 56 a | 55 a | 50 a | 43 a | 57 a |
| SOC *, % | 7.51 d | 2.82 c | 1.31 a | 1.41 ab | 1.46 b |
| SON, % | 0.43 d | 0.11 c | 0.05 b | 0.04 a | 0.03 a |
| SCN | 20 a | 31 b | 29 b | 42 c | 55 d |
| NO3−, mg N kg−1 soil | 2.8 c | 1.0 b | 0.8 a | 0.8 a | 0.8 a |
| NH4+, mg N kg−1 soil | 1.4 b | 0.5 a | 1.6 b | 2.2 c | 1.7 b |
| Labile P2O5, mg kg−1 soil | 1.2 c | 0.5 a | 0.7 b | 0.7 b | 0.5 a |
| STP, mg·kg−1 soil | 85 e | 54 d | 58 c | 53 b | 47 a |
| CO2, μL · hr−1 · g−1 soil | 11.6 c | 5.1 b | 0.7 a | 0.5 a | 0.9 a |
| Index | Soil Genetic Horizon | ||||
|---|---|---|---|---|---|
| A ¥ | AEl | ElBt | Bt | BtC | |
| OTUs richness (S) | 1819 d # | 1525 c | 1341 bc | 1155 ab | 1016 a |
| Chao-1 | 2541 d | 2234 c | 1823 b | 1607 ab | 1359 a |
| Dominance (D) | 0.007 a | 0.009 ab | 0.007 a | 0.14 b | 0.27 c |
| Berger-Parker | 0.52 b | 0.52 b | 0.34 a | 0.73 c | 0.21 d |
| Equitability | 0.83 d | 0.80 c | 0.82 cd | 0.77 ab | 0.73 a |
| Shannon | 6.3 d | 5.8 c | 5.9 c | 5.5 b | 5.1 a |
| Simpson (1-D) | 0.993 c | 0.991 bc | 0.993 c | 0.999 b | 0.973 a |
| Evenness | 0.18 bc | 0.19 bc | 0.21 c | 0.15 ab | 0.12 a |
| Index | Soil Genetic Horizon | ||||
|---|---|---|---|---|---|
| A ¥ | AEl | ElBt | Bt | BtC | |
| OTUs richness | 191 c # | 168 bc | 148 b | 112 ba | 94 a |
| Chao-1 | 260 d | 207 c | 158 b | 131 ab | 106 a |
| Dominance (D) | 0.51 b | 0.14 a | 0.08 a | 0.36 ab | 0.33 ab |
| Berger-Parker | 0.63 b | 0.26 ab | 0.20 a | 0.52 ab | 0.52 ab |
| Equitability | 0.26 a | 0.49 bc | 0.64 c | 0.41 ab | 0.42 ab |
| Shannon | 1.4 a | 2.5 ab | 3.2 b | 1.9 a | 1.9 a |
| Simpson (1-D) | 0.49 a | 0.86 b | 0.92 b | 0.64 ab | 0.67 ab |
| Evenness | 0.17 a | 0.06 b | 0.17 c | 0.07 a | 0.08 ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naumova, N.B.; Belanov, I.P.; Alikina, T.Y.; Kabilov, M.R. Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons. Soil Syst. 2021, 5, 14. https://0-doi-org.brum.beds.ac.uk/10.3390/soilsystems5010014
Naumova NB, Belanov IP, Alikina TY, Kabilov MR. Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons. Soil Systems. 2021; 5(1):14. https://0-doi-org.brum.beds.ac.uk/10.3390/soilsystems5010014
Chicago/Turabian StyleNaumova, Natalia B., Ivan P. Belanov, Tatiana Y. Alikina, and Marsel R. Kabilov. 2021. "Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons" Soil Systems 5, no. 1: 14. https://0-doi-org.brum.beds.ac.uk/10.3390/soilsystems5010014






