The Multifactorial Etiopathogeneses Interplay of Inflammatory Bowel Disease: An Overview
Abstract
:1. Introduction
2. Methods
3. Results
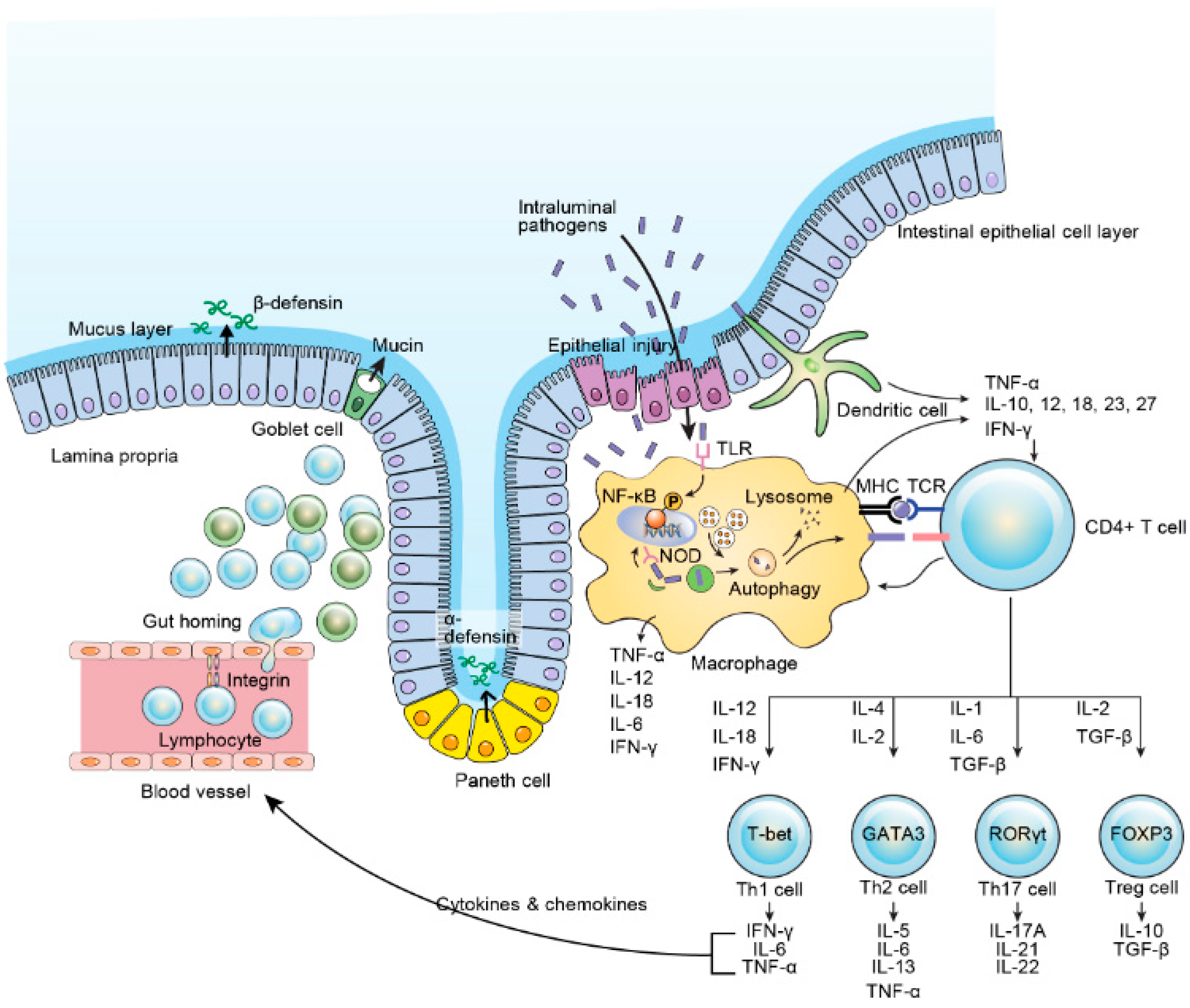
4. Etiopathogenesis of IBD
4.1. Genetic Risk Factors
4.2. Intestinal Microbiota
4.3. The Intestinal Epithelium and Microbiota
4.3.1. Escherichia coli (AIEC)
4.3.2. Clostridium
4.4. Environmental Factors
4.5. Immune Response
4.6. Urbanization
4.7. Dietary/Nutrition
4.8. Role of miRNA in IBD trigger
5. Clinical Diagnosis
6. Management and Challenges
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| IBD | inflammatory bowel Disease |
| UC | ulcerative colitis |
| CD | Crohn’s disease |
| MOOSE | meta-analyses of observational studies |
| PRISMA-P | preferred reporting items for systematic review and meta-analysis protocols |
| MEDLINE | Medical Literature Analysis and retrieval system online |
| EMBASE | Excerpta Medica dataBASE |
| CINAHL | Cumulative index of Nursing and Allied Health Literature |
| SCFAs | short-chain fatty acids |
| NOD2 | Nucleotide-binding oligomerization domain-containing protein 2 |
| TNF-α | tumor necrosis factor-alpha |
References
- Podolsky, D.K. Inflammatory bowel disease. N. Engl. J. Med. 2002, 347, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Love, J.R.; Irvine, E.J.; Fedorak, R.N. Quality of life in inflammatory bowel disease. J. Clin. Gastroenterol. 1992, 14, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Sands, B.E. Therapy of inflammatory bowel disease. Gastroenterology 2000, 118, S68–S82. [Google Scholar] [CrossRef]
- M’Koma, A.E. Inflammatory Bowel Disease: An Expanding Global Health Problem. Clin. Med. Insights Gastroenterol. 2013, 6, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Ventham, N.T.; Kennedy, N.A.; Nimmo, E.R.; Satsangi, J. Beyond gene discovery in inflammatory bowel disease: The emerging role of epigenetics. Gastroenterology 2013, 145, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012, 142, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, A.; Benchimol, E.I.; Bernstein, C.N.; Bitton, A.; Feagan, B.; Panaccione, R.; Glasgow, K.W.; Fernandes, A.; Ghosh, S. Inflammatory bowel disease: A Canadian burden of illness review. Can. J. Gastroenterol. 2012, 26, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Hammer, T.; Nielsen, K.R.; Munkholm, P.; Burisch, J.; Lynge, E. The Faroese IBD Study: Incidence of Inflammatory Bowel Diseases across 54 Years of Population-based Data. J. Crohns Colitis 2016, 10, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Benchimol, E.I.; Manuel, D.G.; Guttmann, A.; Nguyen, G.C.; Mojaverian, N.; Quach, P.; Mack, D.R. Changing age demographics of inflammatory bowel disease in Ontario, Canada: A population-based cohort study of epidemiology trends. Inflamm. Bowel Dis. 2014, 20, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Benchimol, E.I.; Guttmann, A.; Griffiths, A.M.; Rabeneck, L.; Mack, D.R.; Brill, H.; Howard, J.; Guan, J.; To, T. Increasing incidence of paediatric inflammatory bowel disease in Ontario, Canada: Evidence from health administrative data. Gut 2009, 58, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2018, 390, 2769–2778. [Google Scholar] [CrossRef]
- M’Koma, A.E. Diagnosis of inflammatory bowel disease: Potential role of molecular biometrics. World J. Gastrointest. Surg. 2014, 6, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.G.; Ng, S.C. Globalisation of inflammatory bowel disease: Perspectives from the evolution of inflammatory bowel disease in the UK and China. Lancet Gastroenterol. Hepatol. 2016, 1, 307–316. [Google Scholar] [CrossRef]
- Everhov, Å.H.; Halfvarson, J.; Myrelid, P.; Sachs, M.C.; Nordenvall, C.; Söderling, J.; Ekbom, A.; Neovius, M.; Ludvigsson, J.F.; Askling, J.; Olén, O. Incidence and Treatment of Patients Diagnosed With Inflammatory Bowel Diseases at 60 Years or Older in Sweden. Gastroenterology 2018, 154, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Archampong, T.N.; Nkrumah, K.N. Inflammatory bowel disease in Accra: What new trends. West Afr. J. Med. 2013, 32, 40–44. [Google Scholar] [PubMed]
- Ukwenya, A.Y.; Ahmed, A.; Odigie, V.I.; Mohammed, A. Inflammatory bowel disease in Nigerians: Still a rare diagnosis? Ann. Afr. Med. 2011, 10, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Agoda-Koussema, L.K.; Anoukoum, T.; Djibril, A.M.; Balaka, A.; Folligan, K.; Adjenou, V.; Amouzou, K.D.; N’dakéna, K.; Redah, R. Ulcerative colitis: A case in Togo. Med. Sante Trop. 2012, 22, 79–81. [Google Scholar] [PubMed]
- Mebazaa, A.; Aounallah, A.; Naija, N.; Cheikh Rouhou, R.; Kallel, L.; El Euch, D.; Boubaker, J.; Mokni, M.; Filali, A.; Ben Osman, A. Dermatologic manifestations in inflammatory bowel disease in Tunisia. Tunis Med. 2012, 90, 252–257. [Google Scholar] [PubMed]
- Senbanjo, I.O.; Oshikoya, K.A.; Onyekwere, C.A.; Abdulkareem, F.B.; Njokanma, O.F. Ulcerative colitis in a Nigerian girl: A case report. BMC Res. Notes 2012, 5, 564. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, D.; Fourati, H.; Amouri, A.; Marques, I.; Abida, O.; Haddouk, S.; Ben Ayed, M.; Tahri, N.; Penha-Gonçalves, C.; Masmoudi, H. The CREM gene is involved in genetic predisposition to inflammatory bowel disease in the Tunisian population. Hum. Immunol. 2011, 72, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, E.A.; Wright, J.P.; Froggatt, J.; Cuming, L.; Elliot, M. Medium-term follow-up of ulcerative colitis in Cape Town. S. Afr. Med. J. 1989, 76, 142–145. [Google Scholar] [PubMed]
- O’Keefe, E.A.; Wright, J.P.; Froggatt, J.; Zabow, D. Medium-term follow-up of Crohn’s disease in Cape Town. S. Afr. Med. J. 1989, 76, 139–141. [Google Scholar] [PubMed]
- Segal, I. Ulcerative colitis in a developing country of Africa: The Baragwanath experience of the first 46 patients. Int. J. Colorectal Dis. 1988, 3, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Segal, I.; Tim, L.O.; Hamilton, D.G.; Walker, A.R. The rarity of ulcerative colitis in South African blacks. Am. J. Gastroenterol. 1980, 74, 332–336. [Google Scholar] [PubMed]
- Wright, J.P.; Marks, I.N.; Jameson, C.; Garisch, J.A.; Burns, D.G.; Kottler, R.E. Inflammatory bowel disease in Cape Town, 1975–1980. Part II. Crohn’s disease. S. Afr. Med. J. 1983, 63, 226–229. [Google Scholar] [PubMed]
- Wright, J.P.; Marks, I.N.; Jameson, C.; Garisch, J.A.; Burns, D.G.; Kottler, R.E. Inflammatory bowel disease in Cape Town, 1975–1980. Part I. Ulcerative colitis. S. Afr. Med. J. 1983, 63, 223–226. [Google Scholar] [PubMed]
- Brom, B.; Bank, S.; Marks, I.N.; Barbezat, G.O.; Raynham, B. Crohn’s disease in the Cape: A follow-up study of 24 cases and a review of the diagnosis and management. S. Afr. Med. J. 1968, 42, 1099–1107. [Google Scholar] [PubMed]
- Novis, B.H.; Marks, I.N.; Bank, S.; Louw, J.H. Incidence of Crohn’s disease at Groote Schuur Hospital during 1970–1974. S. Afr. Med. J. 1975, 49, 693–697. [Google Scholar] [PubMed]
- Sobel, J.D.; Schamroth, L. Ulcerative colitis in the South African Bantu. Gut 1970, 11, 760–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraud, R.M.; Luke, I.; Schmaman, A. Crohn’s disease in the Transvaal Bantu: A report of 5 cases. S. Afr. Med. J. 1969, 43, 610–613. [Google Scholar] [PubMed]
- Ananthakrishnan, A.N.; Kwon, J.; Raffals, L.; Sands, B.; Stenson, W.F.; McGovern, D.; Kwon, J.H.; Rheaume, R.L.; Sandler, R.S. Variation in treatment of patients with inflammatory bowel diseases at major referral centers in the United States. Clin. Gastroenterol. Hepatol. 2015, 13, 1197–1200. [Google Scholar] [CrossRef] [PubMed]
- Kappelman, M.D.; Rifas-Shiman, S.L.; Porter, C.Q.; Ollendorf, D.A.; Sandler, R.S.; Galanko, J.A.; Finkelstein, J.A. Direct health care costs of Crohn’s disease and ulcerative colitis in US children and adults. Gastroenterology 2008, 135, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Chouraki, V.; Savoye, G.; Dauchet, L.; Vernier-Massouille, G.; Dupas, J.L.; Merle, V.; Laberenne, J.E.; Salomez, J.L.; Lerebours, E.; Turck, D.; et al. The changing pattern of Crohn’s disease incidence in northern France: A continuing increase in the 10- to 19-year-old age bracket (1988–2007). Aliment. Pharmacol. Ther. 2011, 33, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, C.; Paerregaard, A.; Munkholm, P.; Faerk, J.; Lange, A.; Andersen, J.; Jakobsen, M.; Kramer, I.; Czernia-Mazurkiewicz, J.; Wewer, V. Pediatric inflammatory bowel disease: Increasing incidence, decreasing surgery rate, and compromised nutritional status: A prospective population-based cohort study 2007–2009. Inflamm. Bowel Dis. 2011, 17, 2541–2550. [Google Scholar] [CrossRef] [PubMed]
- North American Society for Pediatric Gastroenterology Hepatology, and Nutrition and the Crohn’s Colitis Foundation of America. Differentiating ulcerative colitis from Crohn disease in children and young adults. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 653–674. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.M. Specificities of inflammatory bowel disease in childhood. Best Pract. Res. Clin. Gastroenterol. 2004, 18, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.N.; Shanahan, F. Disorders of a modern lifestyle: Reconciling the epidemiology of inflammatory bowel diseases. Gut 2008, 57, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Rogler, G.; Bernstein, C.N.; Sood, A.; Goh, K.L.; Yamamoto-Furusho, J.K.; Abbas, Z.; Fried, M. Role of biological therapy for inflammatory bowel disease in developing countries. Gut 2012, 61, 706–712. [Google Scholar] [CrossRef] [PubMed]
- M’Koma, A.E.; Wise, P.E.; Muldoon, R.L.; Schwartz, D.A.; Washington, M.K.; Herline, A.J. Evolution of the restorative proctocolectomy and its effects on gastrointestinal hormones. Int. J. Colorectal Dis. 2007, 22, 1143–1163. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Greenland, S. Quantitative methods in the review of epidemiologic literature. Epidemiol. Rev. 1987, 9, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ 2015, 350, g7647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalili, H.; Chan, S.S.M.; Lochhead, P.; Ananthakrishnan, A.N.; Hart, A.R.; Chan, A.T. The role of diet in the aetiopathogenesis of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Goethel, A.; Bedrani, L.; Croitoru Mdcm, K. Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm. Bowel Dis. 2018, 24, 1133–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Cheon, J.H. Pathogenesis of Inflammatory Bowel Disease and Recent Advances in Biologic Therapies. Immune Netw. 2017, 17, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Maloy, K.J.; Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 2011, 474, 298–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cader, M.Z.; Kaser, A. Recent advances in inflammatory bowel disease: Mucosal immune cells in intestinal inflammation. Gut 2013, 62, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef] [PubMed]
- Cheon, J.H. Genetics of inflammatory bowel diseases: A comparison between Western and Eastern perspectives. J. Gastroenterol. Hepatol. 2013, 8, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, K.A.; Targan, S.R. Role of cytokines in the pathogenesis of inflammatory bowel disease. Annu. Rev. Med. 2000, 51, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Song, I.D.; Kim, Y.H.; Koo, J.S.; Kim, Y.S.; Kim, J.S.; Kim, N.; Kim, E.S.; Kim, J.H.; Kim, J.W. Efficacy and Safety of Infliximab Therapy and Predictors of Response in Korean Patients with Crohn’s Disease: A Nationwide, Multicenter Study. Yonsei Med. J. 2016, 57, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Anderson, C.A. Genetic studies of Crohn’s disease: Past, present and future. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Moller, F.T.; Andersen, V.; Wohlfahrt, J.; Jess, T. Familial risk of inflammatory bowel disease: A population-based cohort study 1977–2011. Am. J. Gastroenterol. 2015, 110, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Brant, S.R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology 2011, 140, 1704–1712. [Google Scholar] [CrossRef] [PubMed]
- Desai, H.G.; Gupte, P.A. Increasing incidence of Crohn’s disease in India: Is it related to improved sanitation? Indian J. Gastroenterol. 2005, 24, 23–24. [Google Scholar] [PubMed]
- Zheng, J.J.; Zhu, X.S.; Huangfu, Z.; Gao, Z.X.; Guo, Z.R.; Wang, Z. Crohn’s disease in mainland China: A systematic analysis of 50 years of research. Chin. J. Dig. Dis. 2005, 6, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Tsironi, E.; Feakins, R.M.; Probert, C.S.; Rampton, D.S.; Phil, D. Incidence of inflammatory bowel disease is rising and abdominal tuberculosis is falling in Bangladeshis in East London, United Kingdom. Am. J. Gastroenterol. 2004, 99, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Halme, L.; Paavola-Sakki, P.; Turunen, U.; Lappalainen, M.; Farkkila, M.; Kontula, K. Family and twin studies in inflammatory bowel disease. World J. Gastroenterol. 2006, 12, 3668–3672. [Google Scholar] [CrossRef] [PubMed]
- Thia, K.T.; Loftus, E.V., Jr.; Sandborn, W.J.; Yang, S.K. An update on the epidemiology of inflammatory bowel disease in Asia. Am. J. Gastroenterol. 2008, 103, 3167–3182. [Google Scholar] [CrossRef] [PubMed]
- Levi, Z.; Shamiss, A.; Fraser, G.M.; Furman, M.; Derazne, E.; Tzur, D.; Gordon, B.; Welinsky, S.; Gingold Belfer, R.; Afek, A. The Increasing Prevalence of Inflammatory Bowel Diseases Among Jewish Adolescents and the Sociodemographic Factors Associated with Diagnosis. Inflamm. Bowel. Dis. 2013, 19, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.A.; Avila, B.E.; Koskela, J.; Huang, H.; Stevens, C.; Pirinen, M.; Haritunians, T.; Neale, B.M.; Kurki, M.; Ganna, A. Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 2018, 14, e1007329. [Google Scholar] [CrossRef] [PubMed]
- Nakagome, S.; Mano, S.; Kozlowski, L.; Bujnicki, J.M.; Shibata, H.; Fukumaki, Y.; Kidd, J.R.; Kidd, K.K.; Kawamura, S.; Oota, H. Crohn’s disease risk alleles on the NOD2 locus have been maintained by natural selection on standing variation. Mol. Biol. Evol. 2012, 29, 1569–1585. [Google Scholar] [CrossRef] [PubMed]
- Schurr, E.; Gros, P. A common genetic fingerprint in leprosy and Crohn’s disease? N. Engl. J. Med. 2009, 361, 2666–2668. [Google Scholar] [CrossRef] [PubMed]
- Ostrer, H.; Skorecki, K. The population genetics of the Jewish people. Hum. Genet. 2013, 132, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Moltke, I.; Grarup, N.; Jørgensen, M.E.; Bjerregaard, P.; Treebak, J.T.; Fumagalli, M.; Korneliussen, T.S.; Andersen, M.A.; Nielsen, T.S.; Krarup, N.T. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 2014, 512, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.T.; Würtz, P.; Havulinna, A.S.; Palta, P.; Tukiainen, T.; Rehnström, K.; Esko, T.; Mägi, R.; Inouye, M.; Lappalainen, T. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014, 10, e1004494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuk, O.; Schaffner, S.F.; Samocha, K.; Do, R.; Hechter, E.; Kathiresan, S.; Daly, M.J.; Neale, B.M.; Sunyaev, S.; Lander, E.S. Searching for missing heritability: Designing rare variant association studies. Proc. Natl. Acad. Sci. USA 2014, 111, E455–E464. [Google Scholar] [CrossRef] [PubMed]
- Bahcall, O.; Orli, B. Rare variant association studies. Nat. Genet. 2014, 46, 219. [Google Scholar] [CrossRef]
- Auer, P.L.; Nalls, M.; Meschia, J.F.; Worrall, B.B.; Longstreth, W.T., Jr.; Seshadri, S.; Kooperberg, C.; Burger, K.M.; Carlson, C.S.; Carty, C.L. A genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet. 2012, 8, e1002559. [Google Scholar]
- Karban, A.; Eliakim, R.; Brant, S.R. Genetics of inflammatory bowel disease. Isr. Med. Assoc. J. 2002, 4, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Baskovich, B.; Hiraki, S.; Upadhyay, K.; Meyer, P.; Carmi, S.; Barzilai, N.; Darvasi, A.; Ozelius, L.; Peter, I.; Cho, J.H. Expanded genetic screening panel for the Ashkenazi Jewish population. Genet. Med. 2016, 18, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Kaser, A.; Zeissig, S.; Blumberg, R.S. Inflammatory bowel disease. Annu. Rev. Immunol. 2010, 8, 573–621. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.H.; Hilmi, I.; Lian, L.; Patmanathan, S.N.; Hoe, S.Z.; Lee, W.S.; Goh, K.L. Association between inflammatory bowel disease gene 5 (IBD5) and interleukin-23 receptor (IL23R) genetic polymorphisms in Malaysian patients with Crohn’s disease. J. Dig. Dis. 2012, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Goethel, A.; Croitoru, K.; Philpott, D.J. The interplay between microbes and the immune response in inflammatory bowel disease. J. Physiol. 2018, 596, 3869–3882. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St. Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, D.A.; Frank, D.N.; Pace, N.R.; Gordon, J.I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 2008, 3, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Moran, C.; Shanahan, F. The microbiota in inflammatory bowel disease. J. Gastroenterol. 2015, 50, 495–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Imaeda, H.; Takahashi, K.; Kasumi, E.; Bamba, S.; Fujiyama, Y.; Andoh, A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J. Gastroenterol. Hepatol. 2013, 28, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Imaeda, H.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion 2016, 93, 59–565. [Google Scholar] [CrossRef] [PubMed]
- Varela, E.; Manichanh, C.; Gallart, M.; Torrejón, A.; Borruel, N.; Casellas, F.; Guarner, F.; Antolin, M. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment. Pharmacol. Ther. 2013, 38, 151–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, M.; Dogan, B.; Rishniw, M.; Weitzman, G.; Bosworth, B.; Yantiss, R.; Orsi, R.H.; Wiedmann, M.; McDonough, P.; Kim, S.G.; et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007, 1, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Kotlowski, R.; Bernstein, C.N.; Sepehri, S.; Krause, D.O. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 2007, 56, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.M.; Campbell, B.J.; Hart, C.A.; Mpofu, C.; Nayar, M.; Singh, R.; Englyst, H.; Williams, H.F.; Rhodes, J.M. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004, 127, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Antolin, M.; Santos, J.; Torrejon, A.; Casellas, F.; Borruel, N.; Guarner, F.; Malagelada, J.R. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am. J. Gastroenterol. 2008, 103, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Mylonaki, M.; Rayment, N.B.; Rampton, D.S.; Hudspith, B.N.; Brostoff, J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm. Bowel. Dis. 2005, 11, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Neut, C.; Bulois, P.; Desreumaux, P.; Membré, J.M.; Lederman, E.; Gambiez, L.; Cortot, A.; Quandalle, P.; van Kruiningen, H.; Colombel, J.F. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am. J. Gastroenterol. 2002, 97, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.L.; Barnich, N.; Bringer, M.A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Roy, B.C.; Khan, S.A.; Septer, S.; Umar, S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms 2016, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- van der Waaij, L.A.; Harmsen, H.J.; Madjipour, M.; Kroese, F.G.; Zwiers, M.; van Dullemen, H.M.; de Boer, N.K.; Welling, G.W.; Jansen, P.L. Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA-based fluorescent probes: Commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm. Bowel. Dis. 2005, 11, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Schultsz, C.; Van Den Berg, F.M.; Ten Kate, F.W.; Tytgat, G.N.; Dankert, J. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 1999, 117, 1089–1097. [Google Scholar] [CrossRef]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Loubinoux, J.; Bronowicki, J.P.; Pereira, I.A.; Mougenel, J.L.; Faou, A.E. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 2002, 40, 107–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinkevich, V.V.; Beech, I.B. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol. Ecol. 2000, 34, 147–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Ann. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.; Quera, R. Small intestinal bacterial overgrowth: Roles of antibiotics, prebiotics, and probiotics. Gastroenterology 2006, 130, S78–S90. [Google Scholar] [CrossRef] [PubMed]
- Pickard, K.M.; Bremner, A.R.; Gordon, J.N.; MacDonald, T.T. Microbial-gut interactions in health and disease. Immune responses. Best Pract. Res. Clin. Gastroenterol. 2004, 18, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Nagao-Kitamoto, H.; Kitamoto, S.; Kuffa, P.; Kamada, N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest. Res. 2016, 14, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.J.; Ullman, T.A.; Ford, A.C.; Abreu, M.T.; Abadir, A.; Marshall, J.K.; Talley, N.J.; Moayyedi, P. Antibiotic therapy in inflammatory bowel disease: A systematic review and meta-analysis. Am. J. Gastroenterol. 2011, 106, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Mitsuyama, K.; Niwa, M.; Takedatsu, H.; Yamasaki, H.; Kuwaki, K.; Yoshioka, S.; Yamauchi, R.; Fukunaga, S.; Torimura, T. Antibody markers in the diagnosis of inflammatory bowel disease. World J. Gastroenterol. 2016, 22, 1304–1310. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Duerr, R.H.; Taylor, K.D.; Brant, S.R.; Rioux, J.D.; Silverberg, M.S.; Daly, M.J.; Steinhart, A.H.; Abraham, C.; Regueiro, M.; Griffiths, A.; et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006, 314, 1461–1463. [Google Scholar] [CrossRef] [PubMed]
- Elson, C.O.; Cong, Y.; McCracken, V.J.; Dimmitt, R.A.; Lorenz, R.G.; Weaver, C.T. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol. Rev. 2005, 206, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Kim, M.S.; Kim, E.; Cheon, J.H.; Lee, Y.S.; Kim, Y.; Lee, S.H.; Seo, S.U.; Shin, S.H.; Choi, S.S.; et al. Enteric Viruses Ameliorate Gut Inflammation via Toll-like Receptor 3 and Toll-like Receptor 7-Mediated Interferon-beta Production. Immunity 2016, 44, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Marlicz, W.; Zuba-Surma, E.; Kucia, M.; Blogowski, W.; Starzynska, T.; Ratajczak, M.Z. Various types of stem cells, including a population of very small embryonic-like stem cells, are mobilized into peripheral blood in patients with Crohn’s disease. Inflamm. Bowel Dis. 2012, 18, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Marlicz, W.; Skonieczna-Zydecka, K.; Dabos, K.J.; Loniewski, I.; Koulaouzidis, A. Emerging concepts in non-invasive monitoring of Crohn’s disease. Therap. Adv. Gastroenterol. 2018, 11, 1756284818769076. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Sitaraman, S.V.; Babbin, B.A.; Gerner-Smidt, P.; Ribot, E.M.; Garrett, N.; Alpern, J.A.; Akyildiz, A.; Theiss, A.L.; Nusrat, A.; et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab. Investig. 2007, 87, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille-Michaud, A. Adherent-invasive Escherichia coli: A putative new E. coli pathotype associated with Crohn’s disease. Int. J. Med. Microbiol. 2002, 292, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Medina, M.; Aldeguer, X.; Lopez-Siles, M.; González-Huix, F.; López-Oliu, C.; Dahbi, G.; Blanco, J.E.; Blanco, J.; Garcia-Gil, L.J.; Darfeuille-Michaud, A. Molecular diversity of Escherichia coli in the human gut: New ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.L.; Zheng, L.B.; Kanazawa, Y.; Shih, D.Q. Immunopathology of inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012, 30, 759–795. [Google Scholar] [CrossRef] [PubMed]
- Feuerer, M.; Hill, J.A.; Kretschmer, K.; von Boehmer, H.; Mathis, D.; Benoist, C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc. Natl. Acad. Sci. USA 2010, 107, 5919–5924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, Y.; Feng, T.; Fujihashi, K.; Schoeb, T.R.; Elson, C.O. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2009, 106, 19256–19261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willing, B.; Halfvarson, J.; Dicksved, J.; Rosenquist, M.; Järnerot, G.; Engstrand, L.; Tysk, C.; Jansson, J.K. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Gollwitzer, E.S.; Marsland, B.J. Microbiota abnormalities in inflammatory airway diseases—Potential for therapy. Pharmacol. Ther. 2014, 141, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Bellaguarda, E.; Chang, E.B. IBD and the gut microbiota—From bench to personalized medicine. Curr. Gastroenterol. Rep. 2015, 17, 15. [Google Scholar] [CrossRef] [PubMed]
- Pigneur, B.; Sokol, H. Fecal microbiota transplantation in inflammatory bowel disease: The quest for the holy grail. Mucosal Immunol. 2016, 9, 1360–1365. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, K.A.; Boesmans, L.; Boets, E. Modulating the microbiota in inflammatory bowel diseases: Prebiotics, probiotics or faecal transplantation? Proc. Nutr. Soc. 2014, 73, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W.; et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015, 149, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Rossen, N.G.; Fuentes, S.; van der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.; Duflou, A.; Löwenberg, M.; van den Brink, G.R.; Mathus-Vliegen, E.M.; de Vos, W.M.; et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 2015, 149, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Colman, R.J.; Rubin, D.T. Fecal microbiota transplantation as therapy for inflammatory bowel disease: A systematic review and meta-analysis. J. Crohns Colitis 2014, 8, 1569–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rachmilewitz, D.; Katakura, K.; Karmeli, F.; Hayashi, T.; Reinus, C.; Rudensky, B.; Akira, S.; Takeda, K.; Lee, J.; Takabayashi, K.; et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 2004, 126, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zuo, Z.X.; Mao, A.P. Effect of probiotics on inducing remission and maintaining therapy in ulcerative colitis, Crohn’s disease, and pouchitis: Meta-analysis of randomized controlled trials. Inflamm. Bowel Dis. 2014, 20, 21–35. [Google Scholar] [CrossRef] [PubMed]
- van der Sloot, K.W.J.; Weersma, R.K.; Dijkstra, G.; Alizadeh, B.Z. Development and validation of a web-based questionnaire to identify environmental risk factors for inflammatory bowel disease: The Groningen IBD Environmental Questionnaire (GIEQ). J. Gastroenterol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Molodecky, N.A.; Kaplan, G.G. Environmental risk factors for inflammatory bowel disease. Gastroenterol. Hepatol. 2010, 6, 339–346. [Google Scholar]
- Jess, T.; Riis, L.; Jespersgaard, C.; Hougs, L.; Andersen, P.S.; Orholm, M.K.; Binder, V.; Munkholm, P. Disease concordance, zygosity, and NOD2/CARD15 status: Follow-up of a population-based cohort of Danish twins with inflammatory bowel disease. Am. J. Gastroenterol. 2005, 100, 2486–2492. [Google Scholar] [CrossRef] [PubMed]
- Gaya, D.R.; Russell, R.K.; Nimmo, E.R.; Satsangi, J. New genes in inflammatory bowel disease: Lessons for complex diseases? Lancet 2006, 367, 1271–1284. [Google Scholar] [CrossRef]
- Loftus, E.V., Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 2004, 126, 1504–1517. [Google Scholar] [CrossRef] [PubMed]
- Halfvarson, J.; Jess, T.; Magnuson, A.; Montgomery, S.M.; Orholm, M.; Tysk, C.; Binder, V.; Järnerot, G. Environmental factors in inflammatory bowel disease: A co-twin control study of a Swedish-Danish twin population. Inflamm. Bowel Dis. 2006, 12, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Benchimol, E.I.; Kaplan, G.G.; Otley, A.R.; Nguyen, G.C.; Underwood, F.E.; Guttmann, A.; Jones, J.L.; Potter, B.K.; Catley, C.A.; Nugent, Z.J.; et al. Rural and Urban Residence During Early Life is Associated with Risk of Inflammatory Bowel Disease: A Population-Based Inception and Birth Cohort Study. Am. J. Gastroenterol. 2017, 112, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.G.; Hubbard, J.; Korzenik, J.; Sands, B.E.; Panaccione, R.; Ghosh, S.; Wheeler, A.J.; Villeneuve, P.J. The inflammatory bowel diseases and ambient air pollution: A novel association. Am. J. Gastroenterol. 2010, 105, 2412–2419. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Albenberg, L.; Compher, C.; Baldassano, R.; Piccoli, D.; Lewis, J.D.; Wu, G.D. Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology 2015, 148, 1087–1106. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Bernstein, C.N.; Iliopoulos, D.; Macpherson, A.; Neurath, M.F.; Ali, R.A.R.; Vavricka, S.R.; Fiocchi, C. Environmental triggers in IBD: A review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Mawdsley, J.E.; Rampton, D.S. Psychological stress in IBD: New insights into pathogenic and therapeutic implications. Gut 2005, 54, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Lerebours, E.; Gower-Rousseau, C.; Merle, V.; Brazier, F.; Debeugny, S.; Marti, R.; Salomez, J.L.; Hellot, M.F.; Dupas, J.L.; Colombel, J.F.; et al. Stressful life events as a risk factor for inflammatory bowel disease onset: A population-based case-control study. Am. J. Gastroenterol. 2007, 102, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Probert, C.S.; Jayanthi, V.; Pinder, D.; Wicks, A.C.; Mayberry, J.F. Epidemiological study of ulcerative proctocolitis in Indian migrants and the indigenous population of Leicestershire. Gut 1992, 33, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Sewell, J.L.; Yee, H.F., Jr.; Inadomi, J.M. Hospitalizations are increasing among minority patients with Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 2010, 16, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Rogler, G.; Vavricka, S. Exposome in IBD: Recent insights in environmental factors that influence the onset and course of IBD. Inflamm. Bowel Dis. 2015, 21, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, P.L.; Vegh, Z.; Lovasz, B.D.; David, G.; Pandur, T.; Erdelyi, Z.; Szita, I.; Mester, G.; Balogh, M.; Szipocs, I.; et al. Is current smoking still an important environmental factor in inflammatory bowel diseases? Results from a population-based incident cohort. Inflamm. Bowel Dis. 2013, 19, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, L.M.; Khalili, H.; Chan, A.T.; Richter, J.M.; Bousvaros, A.; Fuchs, C.S. A prospective study of cigarette smoking and the risk of inflammatory bowel disease in women. Am. J. Gastroenterol. 2012, 107, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Kevans, D.; Turpin, W.; Madsen, K.; Meddings, J.; Shestopaloff, K.; Xu, W.; Moreno-Hagelsieb, G.; Griffiths, A.; Silverberg, M.S.; Paterson, A.; et al. Determinants of intestinal permeability in healthy first-degree relatives of individuals with Crohn’s disease. Inflamm. Bowel Dis. 2015, 21, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.N. Review article: Changes in the epidemiology of inflammatory bowel disease-clues for aetiology. Aliment. Pharmacol. Ther. 2017, 46, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Osterman, M.T.; Bewtra, M.; Lewis, J.D. Passive smoking and inflammatory bowel disease: A meta-analysis. Am. J. Gastroenterol. 2008, 103, 2382–2393. [Google Scholar] [CrossRef] [PubMed]
- Chivese, T.; Esterhuizen, T.M.; Basson, A.R.; Watermeyer, G. The Influence of Second-Hand Cigarette Smoke Exposure during Childhood and Active Cigarette Smoking on Crohn’s Disease Phenotype Defined by the Montreal Classification Scheme in a Western Cape Population, South Africa. PLoS ONE 2015, 10, e0139597. [Google Scholar] [CrossRef] [PubMed]
- Mahid, S.S.; Minor, K.S.; Stromberg, A.J.; Galandiuk, S. Active and passive smoking in childhood is related to the development of inflammatory bowel disease. Inflamm. Bowel. Dis. 2007, 13, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Stevens, B.W.; Wilson, R.G.; Russell, C.N.; Cohen, M.A.; Sturgeon, H.C.; Thornton, A.; Giallourakis, C.; Khalili, H.; Nguyen, D.D.; et al. Early life environment and natural history of inflammatory bowel diseases. BMC Gastroenterol. 2014, 14, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Söderholm, J.D.; Yang, P.C.; Ceponis, P.; Vohra, A.; Riddell, R.; Sherman, P.M.; Perdue, M.H. Chronic stress induces mast cell-dependent bacterial adherence and initiates mucosal inflammation in rat intestine. Gastroenterology 2002, 123, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, E.; Sturniolo, G.C.; Puzzolo, D.; Frisina, N.; Fries, W. Effect of stress on the paracellular barrier in the rat ileum. Gut 2002, 51, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Hu, J.; Ghadermarzi, S.; Raza, A.; O’Connell, D.; Xiao, A.; Ayyaz, F.; Zhi, M.; Zhang, Y.; Parekh, N.K.; et al. Smoking and Inflammatory Bowel Disease: A Comparison of China, India, and the USA. Dig. Dis. Sci. 2018, 63, 2703–2713. [Google Scholar] [CrossRef] [PubMed]
- Salih, A.; Widbom, L.; Hultdin, J.; Karling, P. Smoking is associated with risk for developing inflammatory bowel disease including late onset ulcerative colitis: A prospective study. Scand. J. Gastroenterol. 2018, 53, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Matricon, J.; Barnich, N.; Ardid, D. Immunopathogenesis of inflammatory bowel disease. Self Nonself 2010, 1, 299–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, C.T.; Elson, C.O.; Fouser, L.A.; Kolls, J.K. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu. Rev. Pathol. 2013, 8, 477–512. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.P.; Goswami, R. A Decade of Th9 Cells: Role of Th9 Cells in Inflammatory Bowel Disease. Front. Immunol. 2018, 9, 1139. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Raj, J.P. Role of biologics and biosimilars in inflammatory bowel disease: Current trends and future perspectives. J. Inflamm. Res. 2018, 11, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Morrow, T.; Felcone, L.H. Defining the difference: What Makes Biologics Unique. Biotechnol. Healthc. 2004, 1, 24–29. [Google Scholar] [PubMed]
- Chan, H.C.; Ng, S.C. Emerging biologics in inflammatory bowel disease. J. Gastroenterol. 2017, 52, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Seto, K.C.P.S.; Elmqvist, T. Urbanization, Biodiversity and Ecosystem Service: Challenges and Opportunities: A Global Assessment; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Ng, S.C.; Tang, W.; Ching, J.Y.; Wong, M.; Chow, C.M.; Hui, A.J.; Wong, T.C.; Leung, V.K.; Tsang, S.W.; Yu, H.H.; et al. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-pacific Crohn’s and colitis epidemiology study. Gastroenterology 2013, 145, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Bernstein, C.N.; Vatn, M.H.; Lakatos, P.L.; Loftus, E.V., Jr.; Tysk, C.; O’Morain, C.; Moum, B.; Colombel, J.F.; Epidemiology and Natural History Task Force of the International Organization of Inflammatory Bowel Disease (IOIBD). Geographical variability and environmental risk factors in inflammatory bowel disease. Gut 2013, 62, 630–649. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C. Emerging leadership lecture: Inflammatory bowel disease in Asia: Emergence of a “Western” disease. J. Gastroenterol. Hepatol. 2015, 30, 440–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.J.; Kim, W.H.; Cheon, J.H. Clinical characteristics and treatment of inflammatory bowel disease: A comparison of Eastern and Western perspectives. World J. Gastroenterol. 2014, 20, 11525–115237. [Google Scholar] [CrossRef] [PubMed]
- Tozun, N.; Atug, O.; Imeryuz, N.; Hamzaoglu, H.O.; Tiftikci, A.; Parlak, E.; Dagli, U.; Ulker, A.; Hulagu, S.; Akpinar, H.; et al. Clinical characteristics of inflammatory bowel disease in Turkey: A multicenter epidemiologic survey. J. Clin. Gastroenterol. 2009, 43, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Kirsner, J.B. Historical origins of current IBD concepts. World J. Gastroenterol. 2001, 7, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Soon, I.S.; Molodecky, N.A.; Rabi, D.M.; Ghali, W.A.; Barkema, H.W.; Kaplan, G.G. The relationship between urban environment and the inflammatory bowel diseases: A systematic review and meta-analysis. BMC Gastroenterol. 2012, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, I.; Stegen, J.C.; Maldonado-Gómez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The gut microbiota of rural papua new guineans: Composition, diversity patterns, and ecological processes. Cell Rep. 2015, 11, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Kamm, M.A.; Colombel, J.F.; Ng, S.C. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.G.; Puylaert, P.G.P.; Ou, J.; Vipperla, K.; Brouard, F.M.; Ruder, E.H.; Newton, K.; Carbonero, F.; Gaskins, H.R.; de Vos, W.M.; et al. Distinct Microbiotas Are Present in Urban and Rural Native South Africans, and in African Americans. Gastroenterology 2013, 144. [Google Scholar] [CrossRef]
- Benno, Y.; Endo, K.; Mizutani, T.; Namba, Y.; Komori, T.; Mitsuoka, T. Comparison of fecal microflora of elderly persons in rural and urban areas of Japan. Appl. Environ. Microbiol. 1989, 55, 1100–1105. [Google Scholar] [PubMed]
- Zhang, J.; Zheng, Y.; Guo, Z.; Qiao, J.; Gesudu, Q.; Sun, Z.; Huo, D.; Huang, W.; Huo, Q.; Kwok, L.; et al. The diversity of intestinal microbiota of Mongolians living in Inner Mongolia, China. Benef. Microbes 2013, 4, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Tyakht, A.V.; Kostryukova, E.S.; Popenko, A.S.; Belenikin, M.S.; Pavlenko, A.V.; Larin, A.K.; Karpova, I.Y.; Selezneva, O.V.; Semashko, T.A.; Ospanova, E.A.; et al. Human gut microbiota community structures in urban and rural populations in Russia. Nat. Commun. 2013, 4, 2469. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyakht, A.V.; Alexeev, D.G.; Popenko, A.S.; Kostryukova, E.S.; Govorun, V.M. Rural and urban microbiota: To be or not to be? Gut Microbes 2014, 5, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 2017, 17, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Liang, S.; Carlton, E.J.; Jiang, Q.; Wu, J.; Wang, L.; Remais, J.V. Urbanisation and health in China. Lancet 2012, 379, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Ananthakrishnan, A.N.; McGinley, E.L.; Binion, D.G.; Saeian, K. Ambient air pollution correlates with hospitalizations for inflammatory bowel disease: An ecologic analysis. Inflamm. Bowel Dis. 2011, 17, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- To, N.; Gracie, D.J.; Ford, A.C. The Importance of Smoking Cessation in Improving Disease Course in Crohn’s Disease. Am. J. Gastroenterol. 2016, 111, 1198. [Google Scholar] [CrossRef] [PubMed]
- Nunes, T.; Etchevers, M.J.; García-Sánchez, V.; Ginard, D.; Martí, E.; Barreiro-de Acosta, M.; Gomollón, F.; Arroyo, M.; Bastida, G.; Gonzalez, B.; et al. Impact of Smoking Cessation on the Clinical Course of Crohn’s Disease Under Current Therapeutic Algorithms: A Multicenter Prospective Study. Am. J. Gastroenterol. 2016, 111, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.J.; Cosnes, J.; Mansfield, J.C. Review article: Smoking cessation as primary therapy to modify the course of Crohn’s disease. Aliment. Pharmacol. Ther. 2005, 21, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.L.; Hedin, C.R.; Koutsoumpas, A.; Ng, S.C.; McCarthy, N.E.; Prescott, N.J.; Pessoa-Lopes, P.; Mathew, C.G.; Sanderson, J.; Hart, A.L.; et al. Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm. Bowel Dis. 2012, 18, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Van de Wiele, T.; Gallawa, C.M.; Kubachka, K.M.; Creed, J.T.; Basta, N.; Dayton, E.A.; Whitacre, S.; Du Laing, G.; Bradham, K. Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environ. Health Perspect. 2010, 118, 1004–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Wiele, T.; Vanhaecke, L.; Boeckaert, C.; Peru, K.; Headley, J.; Verstraete, W.; Siciliano, S. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ. Health Perspect. 2005, 113, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, K.; Mundandhara, S.; Ghio, A.J.; Madden, M.C. The effects of ambient particulate matter on human alveolar macrophage oxidative and inflammatory responses. J. Toxicol. Environ. Health A 2010, 73, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Van Eeden, S.F.; Tan, W.C.; Suwa, T.; Mukae, H.; Terashima, T.; Fujii, T.; Qui, D.; Vincent, R.; Hogg, J.C. Cytokines involved in the systemic inflammatory response induced by exposure to particulate matter air pollutants (PM(10)). Am. J. Respir. Crit. Care. Med. 2001, 164, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Rickert, D.E.; Butterworth, B.E.; Popp, J.A. Dinitrotoluene: Acute toxicity, oncogenicity, genotoxicity, and metabolism. Crit. Rev. Toxicol. 1984, 13, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Rickert, D.E.; Long, R.M.; Krakowka, S.; Dent, J.G. Metabolism and excretion of 2,4-[14C]Dinitrotoluene in conventional and axenic Fischer-344 rats. Toxicol. Appl. Pharmacol. 1981, 59, 574–579. [Google Scholar] [CrossRef]
- Yim, Y.J.; Seo, J.; Kang, S.I.; Ahn, J.H.; Hur, H.G. Reductive dechlorination of methoxychlor and DDT by human intestinal bacterium Eubacterium limosum under anaerobic conditions. Arch. Environ. Contam. Toxicol. 2008, 54, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Joly, C.; Gay-Quéheillard, J.; Léké, A.; Chardon, K.; Delanaud, S.; Bach, V.; Khorsi-Cauet, H. Impact of chronic exposure to low doses of chlorpyrifos on the intestinal microbiota in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) and in the rat. Environ. Sci. Pollut. Res. Int. 2013, 20, 2726–2734. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, M.; Hassanzadeh, P.; Alaei, S. Cadmium chloride exhibits a profound toxic effect on bacterial microflora of the mice gastrointestinal tract. Hum. Exp. Toxicol. 2011, 30, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Breton, J.; Massart, S.; Vandamme, P.; De Brandt, E.; Pot, B.; Foligne, B. Ecotoxicology inside the gut: Impact of heavy metals on the mouse microbiome. BMC Pharmacol. Toxicol. 2013, 14, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.; Abo, R.P.; Schlieper, K.A.; Graffam, M.E.; Levine, S.; Wishnok, J.S.; Swenberg, J.A.; Tannenbaum, S.R.; Fox, J.G. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: An integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 2014, 122, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Heinze, T.M.; Chen, S.; Cerniglia, C.E.; Chen, H. Anaerobic metabolism of 1-amino-2-naphthol-based azo dyes (Sudan dyes) by human intestinal microflora. Appl. Environ. Microbiol. 2007, 73, 7759–7762. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhao, A.; Xie, G.; Chi, Y.; Zhao, L.; Li, H.; Wang, C.; Bao, Y.; Jia, W.; Luther, M.; et al. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci. Transl. Med. 2013, 5, 172ra22. [Google Scholar] [CrossRef] [PubMed]
- Kish, L.; Hotte, N.; Kaplan, G.G.; Vincent, R.; Tso, R.; Gänzle, M.; Rioux, K.P.; Thiesen, A.; Barkema, H.W.; Wine, E.; et al. Environmental particulate matter induces murine intestinal inflammatory responses and alters the gut microbiome. PLoS ONE 2013, 8, e62220. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.J.; Eum, S.Y.; Rampersaud, E.; Daunert, S.; Abreu, M.T.; Toborek, M. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect. 2013, 121, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A.; et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.L.; Finlay, B.B. The impact of gut microbes in allergic diseases. Curr. Opin. Gastroenterol. 2012, 28, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Gascon, M.; Morales, E.; Sunyer, J.; Vrijheid, M. Effects of persistent organic pollutants on the developing respiratory and immune systems: A systematic review. Environ. Int. 2013, 52, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Menard, S.; Guzylack-Piriou, L.; Leveque, M.; Braniste, V.; Lencina, C.; Naturel, M.; Moussa, L.; Sekkal, S.; Harkat, C.; Gaultier, E.; et al. Food intolerance at adulthood after perinatal exposure to the endocrine disruptor bisphenol A. FASEB J. 2014, 28, 4893–4900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belles-Isles, M.; Ayotte, P.; Dewailly, E.; Weber, J.P.; Roy, R. Cord blood lymphocyte functions in newborns from a remote maritime population exposed to organochlorines and methylmercury. J. Toxicol. Environ. Health A 2002, 65, 165–182. [Google Scholar] [CrossRef] [PubMed]
- MacGillivray, D.M.; Kollmann, T.R. The role of environmental factors in modulating immune responses in early life. Front. Immunol. 2014, 5, 434. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Andersen, V.; Chan, S.; Luben, R.; Khaw, K.T.; Olsen, A.; Tjonneland, A.; Kaaks, R.; Grip, O.; Bergmann, M.M.; Boeing, H.; et al. Fibre intake and the development of inflammatory bowel disease: A European prospective multi-centre cohort study (EPIC-IBD). J. Crohns Colitis 2018, 12, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Opstelten, J.L.; Leenders, M.; Dik, V.K.; Chan, S.S.; van Schaik, F.D.; Khaw, K.T.; Luben, R.; Hallmans, G.; Karling, P.; Lindgren, S.; et al. Dairy Products, Dietary Calcium, and Risk of Inflammatory Bowel Disease: Results From a European Prospective Cohort Investigation. Inflamm. Bowel Dis. 2016, 22, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zamora-Ros, R.; Chan, S.; Cross, A.J.; Ward, H.; Jakszyn, P.; Luben, R.; Opstelten, J.L.; Oldenburg, B.; Hallmans, G.; et al. Dietary Polyphenols in the Aetiology of Crohn’s Disease and Ulcerative Colitis-A Multicenter European Prospective Cohort Study (EPIC). Inflamm. Bowel Dis. 2017, 23, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Tjonneland, A.; Overvad, K.; Bergmann, M.M.; Nagel, G.; Linseisen, J.; Hallmans, G.; Palmqvist, R.; Sjodin, H.; Hagglund, G.; Berglund, G.; et al. Linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: A nested case-control study within a European prospective cohort study. Gut 2009, 58, 1606–1611. [Google Scholar] [PubMed]
- Shim, J.S.; Oh, K.; Kim, H.C. Dietary assessment methods in epidemiologic studies. Epidemiol. Health 2014, 36, e2014009. [Google Scholar] [CrossRef] [PubMed]
- Racine, A.; Carbonnel, F.; Chan, S.S.; Hart, A.R.; Bueno-de-Mesquita, H.B.; Oldenburg, B.; van Schaik, F.D.; Tjønneland, A.; Olsen, A.; Dahm, C.C.; et al. Dietary Patterns and Risk of Inflammatory Bowel Disease in Europe: Results from the EPIC Study. Inflamm. Bowel Dis. 2016, 22, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.F. Impaired inactivation of digestive proteases by deconjugated bilirubin: The possible mechanism for inflammatory bowel disease. Med. Hypotheses 2002, 59, 159–163. [Google Scholar] [CrossRef]
- Qin, X. Etiology of inflammatory bowel disease: A unified hypothesis. World J. Gastroenterol. 2012, 18, 1708–1722. [Google Scholar] [CrossRef] [PubMed]
- Lange, F.T.; Scheurer, M.; Brauch, H.J. Artificial sweeteners—A recently recognized class of emerging environmental contaminants: A review. Anal. Bioanal. Chem. 2012, 403, 2503–2518. [Google Scholar] [CrossRef] [PubMed]
- Khalili, H.; Hakansson, N.; Chan, S.S.; Ludvigsson, J.F.; Olen, O.; Chan, A.T.; Hart, A.R.; Wolk, A. No Association Between Consumption of Sweetened Beverages and Later Risk of Crohn’s Disease or Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005, 6, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leone, V.A.; Cham, C.M.; Chang, E.B. Diet, gut microbes, and genetics in immune function: Can we leverage our current knowledge to achieve better outcomes in inflammatory bowel diseases? Curr. Opin. Immunol. 2014, 31, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Khalili, H.; Malik, S.; Ananthakrishnan, A.N.; Garber, J.J.; Higuchi, L.M.; Joshi, A.; Peloquin, J.; Richter, J.M.; Stewart, K.O.; Curhan, G.C.; et al. Identification and Characterization of a Novel Association between Dietary Potassium and Risk of Crohn’s Disease and Ulcerative Colitis. Front. Immunol. 2016, 7, 554. [Google Scholar] [CrossRef] [PubMed]
- Van Der Sloot, K.W.; Joshi, A.D.; Bellavance, D.R.; Gilpin, K.K.; Stewart, K.O.; Lochhead, P.; Garber, J.J.; Giallourakis, C.; Yajnik, V.; Ananthakrishnan, A.N.; et al. Visceral Adiposity, Genetic Susceptibility, and Risk of Complications Among Individuals with Crohn’s Disease. Inflamm. Bowel Dis. 2017, 23, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Khalili, H.; Song, M.; Higuchi, L.M.; Lochhead, P.; Richter, J.M.; Chan, A.T. Genetic Polymorphisms in Fatty Acid Metabolism Modify the Association Between Dietary n3: N6 Intake and Risk of Ulcerative Colitis: A Prospective Cohort Study. Inflamm. Bowel Dis. 2017, 23, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Costea, I.; Mack, D.R.; Lemaitre, R.N.; Israel, D.; Marcil, V.; Ahmad, A.; Amre, D.K. Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn’s disease. Gastroenterology 2014, 146, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Khalili, H.; de Silva, P.S.; Ananthakrishnan, A.N.; Lochhead, P.; Joshi, A.; Garber, J.J.; Richter, J.R.; Sauk, J.; Chan, A.T. Dietary Iron and Heme Iron Consumption, Genetic Susceptibility, and Risk of Crohn’s Disease and Ulcerative Colitis. Inflamm. Bowel Dis. 2017, 23, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, C.E.; Rose-Zerilli, M.J.; Machado, L.R.; Iriyama, C.; Hollox, E.J.; Cragg, M.S.; Strefford, J.C. Fcγ receptors: Genetic variation, function, and disease. Immunol. Rev. 2015, 268, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.P.; Edberg, J.C.; Kimberly, R.P.; Mangan, E.K.; Bharadwaj, D.; Mold, C.; Du Clos, T.W. C-reactive protein binding to FcgammaRIIa on human monocytes and neutrophils is allele-specific. J. Clin. Investig. 2000, 105, 369–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, M.P.; Mold, C.; Du Clos, T.W. C-reactive protein binding to murine leukocytes requires Fc gamma receptors. J. Immunol. 2000, 164, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, D.; Stein, M.P.; Volzer, M.; Mold, C.; Du Clos, T.W. The major receptor for C-reactive protein on leukocytes is fcgamma receptor II. J. Exp. Med. 1999, 190, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Ivanov, I.I.; Spolski, R.; Min, R.; Shenderov, K.; Egawa, T.; Levy, D.E.; Leonard, W.J.; Littman, D.R. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007, 8, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Fantini, M.C.; Rizzo, A.; Fina, D.; Caruso, R.; Becker, C.; Neurath, M.F.; Macdonald, T.T.; Pallone, F.; Monteleone, G. IL-21 regulates experimental colitis by modulating the balance between Treg and Th17 cells. Eur. J. Immunol. 2007, 37, 3155–3163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Kalla, R.; Ventham, N.T.; Kennedy, N.A.; Quintana, J.F.; Nimmo, E.R.; Buck, A.H.; Satsangi, J. MicroRNAs: New players in IBD. Gut 2015, 64, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.D.; Andersen, R.F.; Christensen, H.; Nathan, T.; Kjeldsen, J.; Madsen, J.S. Circulating microRNAs as biomarkers of adult Crohn’s disease. Eur. J. Gastroenterol. Hepatol. 2015, 27, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.G.; Pekow, J. The emerging role of miRNAs in inflammatory bowel disease: A review. Therap. Adv. Gastroenterol. 2015, 8, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Dalal, S.R.; Kwon, J.H. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol. Hepatol. 2010, 6, 714–722. [Google Scholar]
- Ratajczak, M.Z.; Ratajczak, J. Horizontal transfer of RNA and proteins between cells by extracellular microvesicles: 14 years later. Clin. Transl. Med. 2016, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Ratajczak, D.; Pedziwiatr, D. Extracellular Microvesicles (ExMVs) in Cell to Cell Communication: A Role of Telocytes. Adv. Exp. Med. Biol. 2016, 913, 41–49. [Google Scholar] [PubMed]
- Ratajczak, M.Z.; Ratajczak, J. Extracellular Microvesicles as Game Changers in Better Understanding the Complexity of Cellular Interactions-From Bench to Clinical Applications. Am. J. Med. Sci. 2017, 354, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Tziatzios, G.; Polymeros, D.; Spathis, A.; Triantafyllou, M.; Gkolfakis, P.; Karakitsos, P.; Dimitriadis, G.; Triantafyllou, K. Increased levels of circulating platelet derived microparticles in Crohn’s disease patients. Scand. J. Gastroenterol. 2016, 51, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Swanson, G.; Behara, R.; Braun, R.; Keshavarzian, A. Diagnostic medical radiation in inflammatory bowel disease: How to limit risk and maximize benefit. Inflamm. Bowel Dis. 2013, 19, 2501–2508. [Google Scholar] [CrossRef] [PubMed]
- Cantoro, L.; Di Sabatino, A.; Papi, C.; Margagnoni, G.; Ardizzone, S.; Giuffrida, P.; Giannarelli, D.; Massari, A.; Monterubbianesi, R.; Lenti, M.V.; et al. The Time Course of Diagnostic Delay in Inflammatory Bowel Disease Over the Last Sixty Years: An Italian Multicentre Study. J. Crohns Colitis 2017, 11, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.L.; Stocchi, L.; Duraes, L.; Rencuzogullari, A.; Bennett, A.E.; Remzi, F.H. Long-Term Outcomes in Indeterminate Colitis Patients Undergoing Ileal Pouch-Anal Anastomosis: Function, Quality of Life, and Complications. J. Gastrointest. Surg. 2017, 21, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Tremaine, W.J. Is indeterminate colitis determinable? Curr. Gastroenterol. Rep. 2012, 14, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Tremaine, W.J. Review article: Indeterminate colitis—Definition, diagnosis and management. Aliment. Pharmacol. Ther. 2007, 25, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Conrad, K.; Roggenbuck, D.; Laass, M.W. Diagnosis and classification of ulcerative colitis. Autoimmun. Rev. 2014, 13, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Laass, M.W.; Roggenbuck, D.; Conrad, K. Diagnosis and classification of Crohn’s disease. Autoimmun. Rev. 2014, 13, 467–471. [Google Scholar] [CrossRef] [PubMed]
- M’Koma, A.E.; Seeley, E.H.; Washington, M.K.; Schwartz, D.A.; Muldoon, R.L.; Herline, A.J.; Wise, P.E.; Caprioli, R.M. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm. Bowel Dis. 2011, 17, 875–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeley, E.H.; Washington, M.K.; Caprioli, R.M.; M’Koma, A.E. Proteomic patterns of colonic mucosal tissues delineate Crohn’s colitis and ulcerative colitis. Proteom. Clin. Appl. 2013, 7, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.D.; Korolkova, O.Y.; Sakwe, A.M.; Geiger, T.M.; James, S.D.; Muldoon, R.L.; Herline, A.J.; Goodwin, J.S.; Izban, M.G.; Washington, M.K.; et al. Human alpha defensin 5 is a candidate biomarker to delineate inflammatory bowel disease. PLoS ONE 2017, 12, e0179710. [Google Scholar]
- Shen, B.; Remzi, F.H.; Brzezinski, A.; Lopez, R.; Bennett, A.E.; Lavery, I.C.; Queener, E.; Fazio, V.W. Risk factors for pouch failure in patients with different phenotypes of Crohn’s disease of the pouch. Inflamm. Bowel Dis. 2008, 14, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Remzi, F.H.; Lavery, I.C.; Lashner, B.A.; Fazio, V.W. A proposed classification of ileal pouch disorders and associated complications after restorative proctocolectomy. Clin. Gastroenterol. Hepatol. 2008, 6, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Mehta, F. Report: Economic implications of inflammatory bowel disease and its management. Am. J. Manag. Care 2016, 22, s51–s60. [Google Scholar] [PubMed]
- Long, M.D.; Sands, B.E. What Is the Role of the Inflammatory Bowel Disease Panel in Diagnosis and Treatment? Clin. Gastroenterol. Hepatol. 2018, 16, 618–620. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N.; Cagan, A.; Cai, T.; Gainer, V.S.; Shaw, S.Y.; Savova, G.; Churchill, S.; Karlson, E.W.; Kohane, I.; Liao, K.P.; et al. Comparative Effectiveness of Infliximab and Adalimumab in Crohn’s Disease and Ulcerative Colitis. Inflamm. Bowel Dis. 2016, 22, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Costello, S.P.; Soo, W.; Bryant, R.V.; Jairath, V.; Hart, A.L.; Andrews, J.M. Systematic review with meta-analysis: Faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment. Pharmacol. Ther. 2017, 46, 213–224. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
M’Koma, A.E. The Multifactorial Etiopathogeneses Interplay of Inflammatory Bowel Disease: An Overview. Gastrointest. Disord. 2019, 1, 75-105. https://0-doi-org.brum.beds.ac.uk/10.3390/gidisord1010007
M’Koma AE. The Multifactorial Etiopathogeneses Interplay of Inflammatory Bowel Disease: An Overview. Gastrointestinal Disorders. 2019; 1(1):75-105. https://0-doi-org.brum.beds.ac.uk/10.3390/gidisord1010007
Chicago/Turabian StyleM’Koma, Amosy E. 2019. "The Multifactorial Etiopathogeneses Interplay of Inflammatory Bowel Disease: An Overview" Gastrointestinal Disorders 1, no. 1: 75-105. https://0-doi-org.brum.beds.ac.uk/10.3390/gidisord1010007




