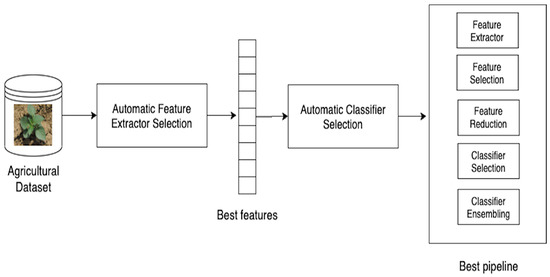
1. Introduction
Nowadays, the damage caused by weeds accounts for important global yield losses and is expected to increase in the coming years [
1]. Although traditionally pesticides were homogeneously applied to solve this problem, there is a tendency in the EU policy to reduce the use of plant protection products since they can cause ground environmental pollution, chemical residues on the crops, and future drug resistance [
2]. More specifically, the EU has set a target to reduce pesticide use by 50% in the next 10 years [
3]. Currently, for applying less dosage of chemical herbicides to weed targets, automatic weed control arises as a possible solution [
4,
5,
6].
Recent advances in image classification techniques provide an opportunity for the improvement of automatic weed control. Despite the delay in the introduction of such techniques to the agricultural domain, the pace that such technologies are being adopted is extremely fast. The use of machine-learning-based image analysis presents a relatively quick, non-invasive, and non-destructive way of controlling weeds spread. In agriculture, deep learning models have been used in the detection of plant diseases and weeds identification [
7,
8,
9,
10]. Convolution Neural Networks (CNNs) are currently the most popular technique in the agricultural domain since, theoretically, they can mitigate some challenges such as inter-class similarities within a plant family and large intra-class variations in background, occlusion, pose, color, and illumination. Besides their good classification performances, some of these works presented deep neural networks whose inference times are suitable for real-time agricultural weed control [
11].
However, there are still challenges to fully adopt the deep learning solutions due to the highly complex agricultural environment, which requires complicated iteratively fine-tuned machine vision algorithms [
12]. Since a suitable machine-learning-based system is the right combination of several components, such as feature extraction, feature selection, and classification, their construction requires knowledge of mathematics, image analysis, coding, and extensive experience in the selection of model architectures [
13,
14]. Therefore, finding the system with the highest performance requires a substantial amount of trial-and-error experimentation time and a highly skilled team to manually test various configurations and models. Additionally, a classifier must be iteratively retrained repeatedly because different conditions can dramatically vary among crops, pest species, areas, and regions. Thus, the ability to automatically recreate a machine learning model specific to each situation, even by non-experts, would be desirable.
To address this situation, Automated Machine Learning (AutoML) systems have arisen in the past years to allow computers to automatically find the most suitable machine learning pipeline matching a specific task and dataset. AutoML systems could provide insights to experienced engineers resulting in better models deployed in a shorter period of time, while allowing inexperienced users to get a glimpse of how such models work, what type of data they require, and how they could be implemented to solve common agricultural problems. AutoML systems are meta-level machine learning algorithms, which use other machine learning solutions as building blocks for finding the optimal ML pipeline structures [
13,
14]. These systems automatically evaluate multiple pipeline configurations, trying to improve the performance iteratively. As a consequence, one of the AutoML systems’ drawbacks is that they consume a lot of computing resources. For that reason, different AutoML cloud-solutions are now offered by IT firms such as Google Cloud AutoML Vision, Microsoft Azure Machine Learning, and Apple’s Create ML. They offer user-friendly interfaces and require little expertise in machine learning to train models. On the other hand, open-source technologies have also arisen to raise awareness of the strengths and limitations of the AutoML systems; for example, AutoKeras, AutoSklearn, Auto-WEKA, H2O AutoML, TPOT, autoxgboost, and OBOE. A summary of these systems can be found in
Table 1.
In the agricultural domain, some recent research studies have made use of the AutoML technique in the past few years, using it to process time series as well as proximal and satellite images. In [
15], the authors tested whether AutoML was a useful tool for the identification of pest insect species by using three aphid species. They constructed models that were trained by photographs of those species under various conditions in Google Cloud AutoML Vision and compared their accuracies of identification. Since the rates of correct identification were over 96% when the models were trained with 400 images per class, they considered AutoML to be useful for pest species identification. In [
16], the author used AutoML through the same platform to classify different types of butterflies, image fruits, and larval host plants. Their average accuracy was around 97.1%. In [
17], AutoML was implemented along with neural network algorithms to classify whether the conditions of rice blast disease were exacerbated or relieved by using five years of climatic data. Although the experiments showed 72% accuracy on average, the model obtained an accuracy of 89% in the exacerbation case. Hence, the effectiveness of the proposed classification model, which combined multiple machine learning models, was confirmed. Finally, an AutoML approach has been applied in [
18], in an attempt to map the Parthenium weed. The authors constructed models by using AutoML technology and 16 other classifiers that were trained by satellite pictures of Sentinel-2 and Landsat 8. AutoML model achieved a higher overall classification accuracy of 88.15% using Sentinel-2 and 74% using Landsat 8, results that confirmed the significance of the AutoML in mapping Parthenium weed infestations using satellite imagery. In [
19], authors used wheat lodging assessment with UAV images for high-throughput plant phenotyping. They compared AutoKeras in image classification and regression tasks to transfer learning techniques.
Although the aforementioned research studies have evaluated AutoML, there is still the need for testing the generalization ability of these techniques with different images taken on the field under real-world conditions. The use of open-source solutions, instead of closed cloud-based ones, is a necessary factor for making the advances more accessible and reproducible. In this paper, the performance of open-source AutoML systems was examined as a tool to speed up and simplify the deployment of machine learning/vision solutions in the agricultural domain. The specific objective of the research was to evaluate whether the integration of AutoML techniques could match, in general, manually-designed architectures. This paper presents three main contributions:
A two-stage methodology integrating AutoML for feature extraction through deep learning and plant identification through classifier ensembles.
An implementation based only on open-source AutoML frameworks and two different publicly available datasets is used for providing transparent and reproducible research.
An analysis of the robustness and overfitting tendency of the AutoML systems on noisy data samples is also presented.
The rest of this paper is organized as follows:
Section 2 explains the methodology proposed and the decisions about the experimental setup.
Section 3 presents the analysis of the results, while
Section 4 is dedicated to discussing the obtained results and the suitability of the methodology. Finally,
Section 5 wraps up the paper with conclusions and future work.
4. Discussion
The results have shown that AutoML can provide classifiers with performances over 90% F
1 score. These results were aligned with other related works [
20,
35,
36] but they were lower than our previous work concerning performance [
37]. However, the disadvantage of our previous work was that obtaining better results took significantly longer for fine-tuning precise nuances of deep neural networks by experts on deep learning, and this process could be avoided or shortened by using AutoML. The main effort of the current work was dedicated to creating a reliable experimental setup for evaluating AutoML performance under different conditions, but not for finding suitable ML pipelines, which was done automatically by the Bayesian optimization of the AutoML frameworks. Moreover, it is important to remark that the pipelines evaluated in this work were constrained in some resources (see
Table 2); which means that with more time or trials for finding the correct configuration, the results could improve in both F
1 score and for robustness against overfitting. Since AutoML works and speeds up many tasks, finding a good balance between manual expert machine learning tuning, AutoML and good performances will be a future open topic for research and discussion.
One of the research questions this work tried to answer was whether integrating an AutoML process on top of a deep neural-based feature extractor could increase the performance over a Softmax classifier. According to the results, it is difficult to provide a final answer. Depending on the dataset and the existence (or not) of noise, different patterns can be observed. According to
Figure 4c, the Softmax classifier was able to obtain the highest performances but it was more unstable; the use of a replacement with a single classifier reported a higher median, lower variance, but it did not reach the highest performance. The ensemble approach obtained an intermediate response having the highest median but higher variance than the single classifier approach. As a conclusion, it could be stated that all these possibilities should be evaluated until new research works enlighten more specific results. Finally, from the different configurations evaluated, only the use of plant segmentation reported repeatedly better performances.
Related to the previous research question, it could be discussed whether there was a relevant pattern in the machine learning pipelines on the top of the neural-based feature extraction. According to the results, the Bayesian optimization has followed different paths leading to different combinations of feature selection, dimensional reduction and classifier tuning. This could mean that a slight difference in the dataset could produce a completely different pipeline, where, for instance, the classifier could be either a decision tree or a random forest without any of them overcoming the other option. This is highly related to the no-free-lunch theorem [
38], which could be even more noticeable within AutoML. Additionally, although tree ensembles have been used to avoid overfitting, the results shown in the tables presented some overfitting, which will reduce the applications where these systems could run safely. It could be concluded that AutoML adds a new complexity layer, where a compromise between interpretability and performances should be established according to the final application and its risks.
Another addressed question was whether the evaluation on a noisy dataset would reduce the performance of systems working accurately on a clean dataset. As
Table 4 and
Table 6 show, the answer has been positive. All systems have reduced their performances when trying to classify images that contain noise. This behavior shows the inability to adapt to possible problems that can occur in the field. On the other hand, these noisy images would be easily classified by a human being, and therefore, it would be necessary to find a way to overcome this limitation. The solution evaluated in this paper has been to use noisy samples during training. However, according to
Table 5 and
Table 7, the responses of every pipeline were quite different. There were pipelines that improved in both clean and noisy datasets, while there were other pipelines that reduced their performance in some cases. It could be discussed that depending on the risks and the types of noises that the weeds identification system could suffer, one approach or another should be implemented.
One of the research decisions made during this work was to implement a solution based on open-source technologies. Although the use of some closed cloud-based solutions such as Google Vision can work as a final solution ready for being deployed in production, having access to the whole code and workflow of the solution is desired for advancing machine learning research in general and the AutoML techniques in specific. Due to the novelty of this research area, understanding the factors and parameters which potentially could lead to a better solution, is important. The results shown along with this paper could provide insights about which hyper-parameters should be studied in the future. For instance, batch size (8) and image size (64 × 64) were set as experimental constants, but increasing them could lead to new relevant results in the domain of precision agriculture. After the fine-tuning effort, once an open suitable AutoML pipeline has been found and reported, it could make sense to share the pipeline through a cloud-based solution (not necessarily closed), which could expose the functionality of predicting new samples, and which could be accessed by automatic control weed systems. In summary, closed cloud-based solutions would make more sense being used in the context of operational applications.
Finally, it can be concluded that AutoML could help the agrotechnology community to easily test machine-learning-based solutions requiring fewer resources to be invested in the implementation part of the solution and dedicating more resources to the domain part of the problem. Moreover, due to the dynamic nature of agriculture, the AutoML pipeline presented in this work could easily create a new model based on new samples, speeding up the deployment of high-performing solutions.
5. Conclusions
In this work, a weeds identification system methodology was evaluated by using an integration of two different AutoML systems. Moreover, two different datasets containing 4 and 13 classes of crops, seedlings, and weeds have been used as benchmarks. The best-evaluated systems under the proposed methodology have shown promising performances between 90% and 93% F
1 score depending on the dataset and the existence (or not) of noisy samples. Although results were aligned to previous AutoML works, the implementation of more resource exhaustive practices, such as increasing the batch size while training, will be examined in the future. Moreover, using new datasets, such as the DeepWeeds dataset (
https://github.com/AlexOlsen/DeepWeeds (accessed on 8 February 2021)), will provide more insights about the real generalization ability of the AutoML technology. Additionally, experiments using noisy samples for testing the robustness of the systems will be extended. On the one hand, smearing noise will be used due to its relation with the movement of the vehicles, which could lead to new insights into the implementation of autonomous vehicles used in precision agriculture. On the other hand, training by using one type of noise and evaluating on a test set with a different type of noise will be checked in order to discern any relation among the different types of noise. Finally, since the use of ensembles of decision trees has not avoided a certain degree of overfitting, new machine learning pipelines will be studied. On the one hand, increasing the number of decision trees inside the ensemble will be evaluated; on the other, a new ensemble technique will be studied: the Super Learner [
39], a model stacking method, where a classifier trains on the top of the predictions provided by the individual trained models. For validating its performance, a more sophisticated approach will be used. Every classifier type will be studied separately, constraining the Bayesian search to a smaller subset, which could lead to a better understandability of the obtained results.