Cardiac Development: A Glimpse on Its Translational Contributions
Abstract
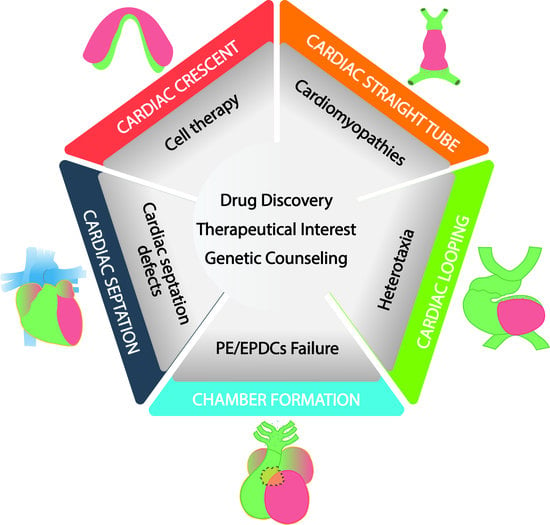
:1. Introduction
1.1. From Gastrulation to the Early Cardiac Linear Tube
1.2. Clinical and Translational Perspectives of Early Cardiogenic Development
1.3. Cardiac Linear Heart and Left–Right Symmetry Break
1.4. Clinical and Translational Perspectives of Left–Right Symmetry Break
1.5. Externally Covering the Naked Myocardium; the Rise of the Epicardium and Its Derivatives
1.6. Clinical and Translational Perspectives of Proepicardium and Epicardium Formation
1.7. Cardiac Ballooning and the Configuration Cardiac Conductive System
1.8. Clinical and Translational Perspectives of Cardiac Ballooning and Conductive Myocardium
1.9. The Formation of a Four-Chambered Heart: Cardiac Septation
1.9.1. Atrial Septation
1.9.2. Atrioventricular Septation
1.9.3. Ventricular Septation
1.9.4. Outflow Tract Septation and Aortic Arch Remodeling
1.10. Clinical and Translational Perspectives of Cardiac Septation
2. Conclusions
3. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
| AAA | Aortic arch anomalies |
| ASD | Atrial septal defects |
| AVC | Atrioventricular canal |
| AVSD | Atrioventricular septal defects |
| Bmp | Bone morphogenetic protein |
| CCS | Cardiac conduction system |
| DORV | Double outlet right ventricle |
| EMT | Epithelial to mesenchymal transition |
| EPDCs | Epicardial derived cells |
| ESC | Embryonic stem cells |
| Fgf | Fibroblast growth factor |
| FHF | First heart field |
| iPS | Induced pluripotent stem cells |
| IVS | Interventricular septum |
| LPM | Lateral plate mesoderm |
| PTA | Permanent truncus arteriosus |
| SHF | Second heart field |
| Shh | Sonic hedghog |
| TGA | Transposition of the great arteries |
| TOF | Tetralogy of Fallot |
| VSD | Ventricular septal defects |
References
- Inman, K.E.; Downs, K.M. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene Expr. Patterns 2006, 6, 783–793. [Google Scholar] [CrossRef]
- Saga, Y.; Miyagawa-Tomita, S.; Takagi, A.; Kitajima, S.; I Miyazaki, J.; Inoue, T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 1999, 126, 3437–3447. [Google Scholar]
- Saga, Y.; Kitajima, S.; Miyagawa-Tomita, S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 2000, 10, 345–352. [Google Scholar] [CrossRef]
- Kitajima, S.; Takagi, A.; Inoue, T.; Saga, Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 2000, 127, 3215–3226. [Google Scholar]
- Bondue, A.; Lapouge, G.; Paulissen, C.; Semeraro, C.; Iacovino, M.; Kyba, M.; Blanpain, C. Mesp1 Acts as a Master Regulator of Multipotent Cardiovascular Progenitor Specification. Cell Stem Cell 2008, 3, 69–84. [Google Scholar] [CrossRef]
- Bondue, A.; Blanpain, C. Mesp1: A key regulator of cardiovascular lineage commitment. Circ. Res. 2010, 107, 1414–1427. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.S.-K.; Shi, X.; Toyama, A.; Arpke, R.W.; Dandapat, A.; Iacovino, M.; Kang, J.-J.; Le, G.; Hagen, H.R.; Garry, D.J.; et al. Mesp1 Patterns Mesoderm into Cardiac, Hematopoietic, or Skeletal Myogenic Progenitors in a Context-Dependent Manner. Cell Stem Cell 2013, 12, 587–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescroart, F.; Chabab, S.; Lin, X.; Rulands, S.; Paulissen, C.; Rodolosse, A.; Auer, H.; Achouri, Y.; Dubois, C.; Bondue, A.; et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 2014, 16, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Chiapparo, G.; Lin, X.; Lescroart, F.; Chabab, S.; Paulissen, C.; Pitisci, L.; Bondue, A.; Blanpain, C. Mesp1 controls the speed, polarity, and directionality of cardiovascular progenitor migration. J. Cell Biol. 2016, 213, 463–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laverriere, A.C.; MacNeill, C.; Mueller, C.; Poelmann, R.E.; Burch, J.B.; Evans, T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994, 269, 23177–23184. [Google Scholar]
- Charron, F.; Nemer, M. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 1999, 10, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Burch, J.B. Regulation of GATA gene expression during vertebrate development. Semin. Cell Dev. Biol. 2005, 16, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Brewer, A.; Pizzey, J. GATA factors in vertebrate heart development and disease. Expert Rev. Mol. Med. 2006, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Evans, T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev. Biol. 1996, 174, 258–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Xu, Q.; Breitbart, R.E. A new tinman-related gene, nkx2.7, anticipate the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev. Biol. 1996, 180, 722–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtzinger, A.; Evans, T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev. Biol. 2007, 312, 613–622. [Google Scholar] [CrossRef] [Green Version]
- Sam, J.; Mercer, E.J.; Torregroza, I.; Banks, K.M.; Evans, T. Specificity, redundancy and dosage thresholds among gata4/5/6 genes during zebrafish cardiogenesis. Biol. Open 2020, 9, bio053611. [Google Scholar] [CrossRef] [PubMed]
- Reecy, J.; Yamada, M.; Cummings, K.; Sosic, D.; Chen, C.-Y.; Eichele, G.; Olson, E.N.; Schwartz, R.J. Chicken Nkx-2.8: A Novel Homeobox Gene Expressed in Early Heart Progenitor Cells and Pharyngeal Pouch-2 and -3 Endoderm. Dev. Biol. 1997, 188, 295–311. [Google Scholar] [CrossRef] [Green Version]
- Boettger, T.; Stein, S.; Kessel, M. The chicken NKX2.8 homeobox gene: A novel member of the NK-2 gene family. Dev. Genes Evol. 1997, 207, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Brand, T.; Andrée, B.; Schneider, A.; Buchberger, A.; Arnold, H.-H. Chicken NKx2-8, a novel homeobox gene expressed during early heart and foregut development. Mech. Dev. 1997, 64, 53–59. [Google Scholar] [CrossRef]
- Jiang, Y.; Tarzami, S.; Burch, J.B.; Evans, T. Common role for each of the cGATA-4/5/6 genes in the regulation of cardiac morphogenesis. Dev. Genet. 1998, 22, 263–277. [Google Scholar] [CrossRef]
- Ban, Q.; Liu, X.; Hui, W.; Chen, D.; Zhao, Z.; Jia, B. Comparative Analysis of Nkx2-5/GATA4/TBX5 Expression in Chicken, Quail and Chicken-quail Hybrids during the Early Stage of Cardiac Development in Embryos. Asian-Australas. J. Anim. Sci. 2013, 26, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Heikinheimo, M.; Scandrett, J.M.; Wilson, D.B. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev. Biol. 1994, 164, 361–373. [Google Scholar] [CrossRef]
- Harvey, R.P. NK-2Homeobox Genes and Heart Development. Dev. Biol. 1996, 178, 203–216. [Google Scholar] [CrossRef] [Green Version]
- Morrisey, E.E.; Ip, H.S.; Lu, M.M.; Parmacek, M.S. GATA-6: A Zinc Finger Transcription Factor That Is Expressed in Multiple Cell Lineages Derived from Lateral Mesoderm. Dev. Biol. 1996, 177, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrisey, E.E.; Ip, H.S.; Tang, Z.; Lu, M.M.; Parmacek, M.S. GATA-5: A Transcriptional Activator Expressed in a Novel Temporally and Spatially-Restricted Pattern during Embryonic Development. Dev. Biol. 1997, 183, 21–36. [Google Scholar] [CrossRef] [Green Version]
- Morrisey, E.E.; Ip, H.S.; Tang, Z.; Parmacek, M.S. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J. Biol. Chem. 1997, 272, 8515–8524. [Google Scholar] [CrossRef] [Green Version]
- Von Both, I.; Silvestri, C.; Erdemir, T.; Lickert, H.; Walls, J.R.; Henkelman, R.M.; Rossant, J.; Harvey, R.P.; Attisano, L.; Wrana, J.L. Foxh1 is essential for development of the anterior heart field. Dev. Cell 2004, 7, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Yamasaki, N.; Izumo, S. Phenotypic Characterization of the Murine Nkx2.6 Homeobox Gene by Gene Targeting. Mol. Cell. Biol. 2000, 20, 2874–2879. [Google Scholar] [CrossRef] [Green Version]
- Caprioli, A.; Koyano-Nakagawa, N.; Iacovino, M.; Shi, X.; Ferdous, A.; Harvey, R.P.; Olson, E.N.; Kyba, M.; Garry, D.J. Nkx2-5 Represses Gata1 Gene Expression and Modulates the Cellular Fate of Cardiac Progenitors During Embryogenesis. Circulation 2011, 123, 1633–1641. [Google Scholar] [CrossRef] [Green Version]
- Kasahara, H.; Bartunkova, S.; Schinke, M.; Tanaka, M.; Izumo, S. Cardiac and Extracardiac Expression of Csx/Nkx2.5 Homeodomain Protein. Circ. Res. 1998, 82, 936–946. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Chen, Z.; Bartunkova, S.; Yamasaki, N.; Izumo, S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 1999, 126, 1269–1280. [Google Scholar] [PubMed]
- Jamali, M.; Rogerson, P.J.; Wilton, S.; Skerjanc, I.S. Nkx2–5 Activity Is Essential for Cardiomyogenesis. J. Biol. Chem. 2001, 276, 42252–42258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, R.P.; Lai, D.; Elliott, D.A.; Biben, C.; Solloway, M.; Prall, O.; Stennard, F.; Schindeler, A.; Groves, N.; Lavulo, L.; et al. Homeodomain Factor Nkx2-5 in Heart Development and Disease. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 107–114. [Google Scholar] [CrossRef]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of Mouse Cardiac Morphogenesis and Myogenesis by Transcription Factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, W.; Drake, C.J.; Schwarz, J.J. The Transcription Factor MEF2C-Null Mouse Exhibits Complex Vascular Malformations and Reduced Cardiac Expression of Angiopoietin 1 and VEGF. Dev. Biol. 1999, 211, 255–267. [Google Scholar] [CrossRef] [Green Version]
- Karamboulas, C.; Dakubo, G.D.; Liu, J.; De Repentigny, Y.; Yutzey, K.; Wallace, V.A.; Kothary, R.; Skerjanc, I.S. Disruption of MEF2 activity in cardiomyoblasts inhibits cardiomyogenesis. J. Cell. Sci. 2006, 119 Pt 20, 4315–4321. [Google Scholar] [CrossRef] [Green Version]
- Materna, S.C.; Sinha, T.; Barnes, R.M.; Van Bueren, K.L.; Black, B.L. Cardiovascular development and survival require Mef2c function in the myocardial but not the endothelial lineage. Dev. Biol. 2019, 445, 170–177. [Google Scholar] [CrossRef]
- Gajewski, K.; Fossett, N.; Molkentin, J.D.; A Schulz, R. The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 1999, 126, 5679–5688. [Google Scholar]
- Pu, W.T.; Ishiwata, T.; Juraszek, A.L.; Ma, Q.; Izumo, S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 2004, 275, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Zhao, R.; Watt, A.J.; Battle, M.A.; Li, J.; Bondow, B.J.; Duncan, S.A. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev. Biol. 2008, 317, 614–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haworth, K.E.; Kotecha, S.; Mohun, T.J.; Latinkić, B.V. GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. BMC Dev. Biol. 2008, 8, 74. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.; Afouda, B.A.; Hoppler, S. Wnt/beta-catenin signalling regulates cardiomyogenesis via GATA transcription factors. J. Anat. 2010, 216, 92–107. [Google Scholar] [CrossRef]
- Clement, J.H.; Fettes, P.; Knöchel, S.; Lef, J.; Knöchel, W. Bone morphogenetic protein 2 in the early development of Xenopus laevis. Mech. Dev. 1995, 52, 357–370. [Google Scholar] [CrossRef]
- Ladd, A.N.; Yatskievych, T.A.; Antin, P.B. Regulation of avian cardiac myogenesis by activin/TGFbeta and bone morphogenetic proteins. Dev. Biol. 1998, 204, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monzen, K.; Shiojima, I.; Hiroi, Y.; Kudoh, S.; Oka, T.; Takimoto, E.; Hayashi, D.; Hosoda, T.; Habara-Ohkubo, A.; Nakaoka, T.; et al. Bone Morphogenetic Proteins Induce Cardiomyocyte Differentiation through the Mitogen-Activated Protein Kinase Kinase Kinase TAK1 and Cardiac Transcription Factors Csx/Nkx-2.5 and GATA-4. Mol. Cell. Biol. 1999, 19, 7096–7105. [Google Scholar] [CrossRef] [Green Version]
- Schlange, T.; Andrée, B.; Arnold, H.-H.; Brand, T. BMP2 is required for early heart development during a distinct time period. Mech. Dev. 2000, 91, 259–270. [Google Scholar] [CrossRef]
- Christiaen, L.A.; Stolfi, A.; Levine, M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 2010, 340, 179–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavrilov, S.; Lacy, E. Genetic dissection of ventral folding morphogenesis in mouse: Embryonic visceral endoderm-supplied BMP2 positions head and heart. Curr. Opin. Genet. Dev. 2013, 23, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Reifers, F.; Walsh, E.C.; Léger, S.; Stainier, D.Y.; Brand, M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar). Development 2000, 127, 225–235. [Google Scholar]
- Lopez-Sanchez, C.; Climent, V.; Schoenwolf, G.C.; Alvarez, I.S.; Garcia-Martinez, V. Induction of cardiogenesis by Hensen’s node and fibroblast growth factors. Cell Tissue Res. 2002, 309, 237–249. [Google Scholar] [CrossRef]
- Alsan, B.H.; Schultheiss, T.M. Regulation of avian cardiogenesis by Fgf8 signaling. Development 2002, 129, 1935–1943. [Google Scholar]
- Ilagan, R.; Abu-Issa, R.; Brown, D.; Yang, Y.-P.; Jiao, K.; Schwartz, R.J.; Klingensmith, J.; Meyers, E.N. Fgf8 is required for anterior heart field development. Development 2006, 133, 2435–2445. [Google Scholar] [CrossRef] [Green Version]
- Marvin, M.J.; Di Rocco, G.; Gardiner, A.; Bush, S.M.; Lassar, A.B. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001, 15, 316–327. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Sano, M.; Songyang, Z.; Schneider, M.D. A Wnt–and beta-catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 5834–5839. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, L.M.; Eisenberg, C.A. Evaluating the role of Wnt signal transduction in promoting the development of the heart. Sci. World J. 2007, 7, 161–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foley, A.C.; Mercola, M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev. 2005, 19, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Klaus, A.; Saga, Y.; Taketo, M.M.; Tzahor, E.; Birchmeier, W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 18531–18536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, T.; Liu, Y.; Jia, Z.; Li, Y.; Zhang, C.; Chen, P.; Ma, K.; Affara, N.; Zhou, C. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim. Biophys. Acta (BBA)–Bioenerg. 2009, 1793, 300–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, R.; Li, D.; Gupta, M.; Manderfield, L.J.; Ifkovits, J.L.; Wang, Q.; Liu, F.; Liu, Y.; Poleshko, A.; Padmanabhan, A.; et al. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science 2015, 348, aaa6071. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Sanchez, C.; Franco, D.; Bonet, F.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V. Negative Fgf8-Bmp2 feed-back is regulated by miR-130 during early cardiac specification. Dev. Biol. 2015, 406, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Christoffels, V.M.; Habets, P.E.; Franco, D.; Campione, M.; de Jong, F.; Lamers, W.H.; Bao, Z.Z.; Palmer, S.; Biben, C.; Harvey, R.P.; et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000, 223, 266–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meilhac, S.M.; Kelly, R.G.; Rocancourt, D.; Eloy-Trinquet, S.; Nicolas, J.-F.; Buckingham, M.E. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development 2003, 130, 3877–3889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meilhac, S.M.; Esner, M.; Kelly, R.G.; Nicolas, J.-F.; Buckingham, M.E. The Clonal Origin of Myocardial Cells in Different Regions of the Embryonic Mouse Heart. Dev. Cell 2004, 6, 685–698. [Google Scholar] [CrossRef] [Green Version]
- Buckingham, M.E.; Meilhac, S.M.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835. [Google Scholar] [CrossRef]
- Meilhac, S.M.; Lescroart, F.; Blanpain, C.; Buckingham, M.E. Cardiac cell lineages that form the heart. Cold Spring Harb. Perspect. Med. 2014, 4, a013888. [Google Scholar] [CrossRef] [Green Version]
- Meilhac, S.M.; Buckingham, M. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018, 15, 705–724. [Google Scholar] [CrossRef]
- Zhang, L.; Nomura-Kitabayashi, A.; Sultana, N.; Cai, W.; Cai, X.; Moon, A.M.; Cai, C.-L. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev. Biol. 2014, 390, 68–79. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S.M. Isl1 Identifies a Cardiac Progenitor Population that Proliferates Prior to Differentiation and Contributes a Majority of Cells to the Heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef] [Green Version]
- Dyer, L.A.; Makadia, F.A.; Scott, A.; Pegram, K.; Hutson, M.R.; Kirby, M.L. BMP signaling modulates hedgehog-induced secondary heart field proliferation. Dev. Biol. 2010, 348, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Hinits, Y.; Pan, L.; Walker, C.; Dowd, J.; Moens, C.B.; Hughes, S.M. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 2012, 369, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Van Oort, R.J.; van Rooij, E.; Bourajjaj, M.; Schimmel, J.; Jansen, M.A.; van der Nagel, R.; Doevendans, P.A.; Schneider, M.D.; van Echteld, C.J.; De Windt, L.J. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation 2006, 114, 298–308. [Google Scholar] [CrossRef] [Green Version]
- Oka, T.; Maillet, M.; Watt, A.J.; Schwartz, R.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. Cardiac-Specific Deletion of Gata4 Reveals Its Requirement for Hypertrophy, Compensation, and Myocyte Viability. Circ. Res. 2006, 98, 837–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, J.P.; Collao, A.; Chiong, M.; Maldonado, C.; Adasme, T.; Carrasco, L.; Ocaranza, P.; Bravo-Sagua, R.; González, L.; Díaz-Araya, G.; et al. The transcription factor MEF2C mediates cardiomyocyte hypertrophy induced by IGF-1 signaling. Biochem. Biophys. Res. Commun. 2009, 388, 155–160. [Google Scholar] [CrossRef]
- Liang, Q.; De Windt, L.J.; Witt, S.A.; Kimball, T.R.; Markham, B.E.; Molkentin, J.D. The Transcription Factors GATA4 and GATA6 Regulate Cardiomyocyte Hypertrophy in Vitro and in Vivo. J. Biol. Chem. 2001, 276, 30245–30253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kontaraki, J.E.; Parthenakis, F.I.; Patrianakos, A.; Karalis, I.K.; Vardas, P.E. Altered expression of early cardiac marker genes in circulating cells of patients with hypertrophic cardiomyopathy. Cardiovasc. Pathol. 2007, 16, 329–335. [Google Scholar] [CrossRef]
- Van Berlo, J.H.; Elrod, J.W.; van den Hoogenhof, M.M.; York, A.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ Res. 2010, 107, 1032–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coppola, A.; Romito, A.; Borel, C.; Gehrig, C.; Gagnebin, M.; Falconnet, E.; Izzo, A.; Altucci, L.; Banfi, S.; Antonarakis, S.E.; et al. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014, 12, 323–337. [Google Scholar] [CrossRef]
- Ménard, C.; Grey, C.; Méry, A.; Zeineddine, D.; Aimond, F.; Pucéat, M. Cardiac specification of embryonic stem cells. J. Cell. Biochem. 2004, 93, 681–687. [Google Scholar] [CrossRef]
- Glass, C.; Singla, R.; Arora, A.; Singla, D.K. Mouse Embryonic Stem Cell-Derived Cardiac Myocytes in a Cell Culture Dish. Methods Mol. Biol. 2015, 1299, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takahashi, T.; Esaki, M.; Ushikoshi, H.; Nagano, S.; Fujiwara, H.; Kosai, K. Efficient cardiomyogenic differentiation of embryonic stem cell by fibroblast growth factor 2 and bone morphogenetic protein 2. Circ. J. 2004, 68, 691–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, W.-H. Embryonic and embryonic-like stem cells in heart muscle engineering. J. Mol. Cell. Cardiol. 2011, 50, 320–326. [Google Scholar] [CrossRef]
- Kinney, M.A.; Sargent, C.Y.; McDevitt, T.C. Temporal Modulation of β-Catenin Signaling by Multicellular Aggregation Kinetics Impacts Embryonic Stem Cell Cardiomyogenesis. Stem Cells Dev. 2013, 22, 2665–2677. [Google Scholar] [CrossRef] [Green Version]
- Maltsev, V.A.; Rohwedel, J.; Hescheler, J.; Wobus, A.M. Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech. Dev. 1993, 44, 41–50. [Google Scholar] [CrossRef]
- Paige, S.L.; Plonowska, K.; Xu, A.; Wu, S.M. Molecular regulation of cardiomyocyte differentiation. Circ. Res. 2015, 116, 341–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sluijter, J.P.; Van Mil, A.; Van Vliet, P.; Metz, C.H.; Liu, J.; Doevendans, P.A.; Goumans, M.-J. MicroRNA-1 and -499 Regulate Differentiation and Proliferation in Human-Derived Cardiomyocyte Progenitor Cells. Arter. Thromb. Vasc. Biol. 2010, 30, 859–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turbendian, H.K.; Gordillo, M.; Tsai, S.-Y.; Lu, J.; Kang, G.; Liu, T.-C.; Tang, A.; Liu, S.; Fishman, G.I.; Evans, T. GATA factors efficiently direct cardiac fate from embryonic stem cells. Development 2013, 140, 1639–1644. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhang, A.D.; Kim, L.J.; Nam, Y.-J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Schweizer, P.A.; Darche, F.F.; Ullrich, N.D.; Geschwill, P.; Greber, B.; Rivinius, R.; Seyler, C.; Müller-Decker, K.; Draguhn, A.; Utikal, J.; et al. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res. Ther. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempf, H.; Zweigerdt, R. Scalable Cardiac Differentiation of Pluripotent Stem Cells Using Specific Growth Factors and Small Molecules. Adv. Biochem. Eng. Biotechnol. 2017, 163, 39–69. [Google Scholar] [CrossRef]
- Hartung, S.; Schwanke, K.; Haase, A.; David, R.; Franz, W.-M.; Martin, U.; Zweigerdt, R. Directing Cardiomyogenic Differentiation of Human Pluripotent Stem Cells by Plasmid-Based Transient Overexpression of Cardiac Transcription Factors. Stem Cells Dev. 2013, 22, 1112–1125. [Google Scholar] [CrossRef] [Green Version]
- Ieda, M.; Fu, J.-D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, H.; Ma, Z.; Feng, J.; Gao, P.; Dong, H.; Zhang, Z. Efficient cardiomyogenic differentiation of bone marrow mesenchymal stromal cells by combination of Wnt11 and bone morphogenetic protein 2. Exp. Biol. Med. 2012, 237, 768–776. [Google Scholar] [CrossRef]
- Kuo, C.T.; E Morrisey, E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef] [Green Version]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef] [Green Version]
- Biben, C.; Harvey, R.P. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997, 11, 1357–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997, 16, 5687–5696. [Google Scholar] [CrossRef] [Green Version]
- Skerjanc, I.S.; Petropoulos, H.; Ridgeway, A.G.; Wilton, S. Myocyte enhancer factor 2C and Nkx2-5 up-regulate each other’s expression and initiate cardiomyogenesis in P19 cells. J. Biol. Chem. 1998, 273, 34904–34910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiojima, I.; Komuro, I.; Oka, T.; Hiroi, Y.; Mizuno, T.; Takimoto, E.; Monzen, K.; Aikawa, R.; Akazawa, H.; Yamazaki, T.; et al. Context-dependent Transcriptional Cooperation Mediated by Cardiac Transcription Factors Csx/Nkx-2.5 and GATA-4. J. Biol. Chem. 1999, 274, 8231–8239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincentz, J.W.; Barnes, R.M.; Firulli, B.A.; Conway, S.J.; Firulli, A.B. Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development. Dev. Dyn. 2008, 237, 3809–3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, T.K.; Song, F.F.; Packham, E.A.; Buxton, S.; Robinson, T.E.; Ronksley, J.; Self, T.; Bonser, A.J.; Brook, J.D. Physical Interaction between TBX5 and MEF2C Is Required for Early Heart Development. Mol. Cell. Biol. 2009, 29, 2205–2218. [Google Scholar] [CrossRef] [Green Version]
- Hiroi, Y.; Kudoh, S.; Monzen, K.; Ikeda, Y.; Yazaki, Y.; Nagai, R.; Komuro, I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001, 28, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Maitra, M.; Schluterman, M.K.; Nichols, H.A.; Richardson, J.A.; Lo, C.W.; Srivastava, D.; Garg, V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev. Biol. 2009, 326, 368–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradhan, L.; Gopal, S.; Li, S.; Ashur, S.; Suryanarayanan, S.; Kasahara, H.; Nam, H.-J. Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5. Biochemistry 2016, 55, 1702–1710. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Li, Y.; Li, S.; Cobb, R.M.; Zhou, D.; Lu, M.M.; Epstein, J.A.; Morrisey, E.E.; Gruber, P.J. Gata4 and Gata5 Cooperatively Regulate Cardiac Myocyte Proliferation in Mice. J. Biol. Chem. 2010, 285, 1765–1772. [Google Scholar] [CrossRef] [Green Version]
- Charron, F.; Paradis, P.; Bronchain, O.; Nemer, G.; Nemer, M. Cooperative Interaction between GATA-4 and GATA-6 Regulates Myocardial Gene Expression. Mol. Cell. Biol. 1999, 19, 4355–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prall, O.W.; Menon, M.K.; Solloway, M.J.; Watanabe, Y.; Zaffran, S.; Bajolle, F.; Biben, C.; McBride, J.J.; Robertson, B.R.; Chaulet, H.; et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 2007, 128, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Schlesinger, J.; Schueler, M.; Grunert, M.; Fischer, J.J.; Zhang, Q.; Krueger, T.; Lange, M.; Tönjes, M.; Dunkel, I.; Sperling, S. The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs. PLoS Genet. 2011, 7, e1001313. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Y.; Lu, L.; Lu, X.; Dixon, R.A. Cardiac Gene Activation Analysis in Mammalian Non-Myoblasic Cells by Nkx2-5, Tbx5, Gata4 and Myocd. PLoS ONE 2012, 7, e48028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.R.; McKinsey, T.A.; Xu, H.; Wang, D.Z.; Richardson, J.A.; Olson, E.N. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 1999, 19, 4495–4502. [Google Scholar] [CrossRef] [Green Version]
- Akazawa, H.; Kudoh, S.; Mochizuki, N.; Takekoshi, N.; Takano, H.; Nagai, T.; Komuro, I. A novel LIM protein Cal promotes cardiac differentiation by association with CSX/NKX2-5. J. Cell Biol. 2004, 164, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voronova, A.; Al Madhoun, A.; Fischer, A.; Shelton, M.; Karamboulas, C.; Skerjanc, I.S. Gli2 and MEF2C activate each other’s expression and function synergistically during cardiomyogenesis in vitro. Nucleic Acids Res. 2012, 40, 4723–4724. [Google Scholar] [CrossRef] [Green Version]
- Behrens, A.N.; Iacovino, M.; Lohr, J.L.; Ren, Y.; Zierold, C.; Harvey, R.P.; Kyba, M.; Garry, D.J.; Martin, C.M. Nkx2-5 Mediates Differential Cardiac Differentiation Through Interaction with Hoxa10. Stem Cells Dev. 2013, 22, 2211–2220. [Google Scholar] [CrossRef] [Green Version]
- Clark, C.D.; Lee, K.-H. Second heart field-specific expression of Nkx2-5 requires promoter proximal interaction with Srf. Mech. Dev. 2020, 162, 103615. [Google Scholar] [CrossRef]
- Alexandrovich, A.; Arno, M.; Patient, R.K.; Shah, A.M.; Pizzey, J.A.; Brewer, A.C. Wnt2 is a direct downstream target of GATA6 during early cardiogenesis. Mech. Dev. 2006, 123, 297–311. [Google Scholar] [CrossRef]
- Behrens, A.N.; Ren, Y.; Ferdous, A.; Garry, D.J.; Martin, C.M. Nkx2-5 Regulates Tdgf1 (Cripto) Early During Cardiac Development. J. Clin. Exp. Cardiol. 2013, 1, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Cambier, L.; Plate, M.; Sucov, H.M.; Pashmforoush, M. Nkx2-5 regulates cardiac growth through modulation of Wnt signaling by R-spondin3. Development 2014, 141, 2959–2971. [Google Scholar] [CrossRef] [Green Version]
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.-M.; Black, B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 2004, 131, 3931–3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorn, T.; Goedel, A.; Lam, J.T.; Haas, J.; Tian, Q.; Herrmann, F.; Bundschu, K.; Dobreva, G.; Schiemann, M.; Dirschinger, R.; et al. Direct Nkx2-5 Transcriptional Repression of Isl1 Controls Cardiomyocyte Subtype Identity. Stem Cells 2015, 33, 1113–1129. [Google Scholar] [CrossRef]
- Anderson, D.J.; Kaplan, D.I.; Bell, K.M.; Koutsis, K.; Haynes, J.M.; Mills, R.J.; Phelan, D.G.; Qian, E.L.; Leitoguinho, A.R.; Arasaratnam, D.; et al. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Horton, A.J.; Brooker, J.; Streitfeld, W.S.; Flessa, M.E.; Pillai, B.; Simpson, R.; Clark, C.D.; Gooz, M.B.; Sutton, K.K.; Foley, A.C.; et al. Nkx2–5 Second Heart Field Target Gene Ccdc117 Regulates DNA Metabolism and Proliferation. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-P.; Nakagawa, O.; Nakagawa, M.; Yanagisawa, H.; Passier, R.; Richardson, J.A.; Srivastava, D.; Olson, E.N. CHAMP, A Novel Cardiac-Specific Helicase Regulated by MEF2C. Dev. Biol. 2001, 234, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Wythe, J.D.; Liu, J.; Cartry, J.; Vogler, G.; Mohapatra, B.; Otway, R.T.; Huang, Y.; King, I.N.; Maillet, M.; et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J. Cell Biol. 2011, 193, 1181–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Gong, M.; Wang, Y.; Millard, R.W.; Pasha, Z.; Yang, Y.; Ashraf, M.; Xu, M. Cardiomyocyte Protection by GATA-4 Gene Engineered Mesenchymal Stem Cells Is Partially Mediated by Translocation of miR-221 in Microvesicles. PLoS ONE 2013, 8, e73304. [Google Scholar] [CrossRef] [Green Version]
- Watt, A.J.; Battle, M.A.; Li, J.; Duncan, S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 12573–12578. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Von Gise, A.; Ma, Q.; Rivera-Feliciano, J.; Pu, W.T. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem. Biophys. Res. Commun. 2008, 375, 450–453. [Google Scholar] [CrossRef] [Green Version]
- Zeisberg, E.M.; Ma, Q.; Juraszek, A.L.; Moses, K.; Schwartz, R.J.; Izumo, S.; Pu, W.T. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J. Clin. Investig. 2005, 115, 1522–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moskowitz, I.P.; Kim, J.B.; Moore, M.L.; Wolf, C.M.; Peterson, M.A.; Shendure, J.; Nóbrega, M.A.; Yokota, Y.; Berul, C.; Izumo, S.; et al. A Molecular Pathway Including Id2, Tbx5, and Nkx2-5 Required for Cardiac Conduction System Development. Cell 2007, 129, 1365–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza-Lewis, R.A.; Liu, H.; Sun, C.; Chen, C.; Jiao, K.; Chen, Y.-P. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 2011, 356, 359–369. [Google Scholar] [CrossRef] [Green Version]
- Jay, P.Y.; Harris, B.S.; Maguire, C.T.; Buerger, A.; Wakimoto, H.; Tanaka, M.; Kupershmidt, S.; Roden, D.M.; Schultheiss, T.M.; O’Brien, T.X.; et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J. Clin. Investig. 2004, 113, 1130–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meysen, S.; Marger, L.; Hewett, K.W.; Jarry-Guichard, T.; Agarkova, I.; Chauvin, J.P.; Perriard, J.C.; Izumo, S.; Gourdie, R.G.; Mangoni, M.E.; et al. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev. Biol. 2007, 303, 740–753. [Google Scholar] [CrossRef] [Green Version]
- Harris, B.S.; Spruill, L.; Edmonson, A.M.; Rackley, M.S.; Benson, D.W.; O’Brien, T.X.; Gourdie, R.G. Differentiation of cardiac Purkinje fibers requires precise spatiotemporal regulation of Nkx2-5 expression. Dev. Dyn. 2005, 235, 38–49. [Google Scholar] [CrossRef] [Green Version]
- Biben, C.; Weber, R.; Kesteven, S.; Stanley, E.; McDonald, L.; Elliott, D.A.; Barnett, L.; Köentgen, F.; Robb, L.; Feneley, M.; et al. Cardiac Septal and Valvular Dysmorphogenesis in Mice Heterozygous for Mutations in the Homeobox GeneNkx2-5. Circ. Res. 2000, 87, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Berul, C.; Ishii, M.; Jay, P.; Wakimoto, H.; Douglas, P.; Yamasaki, N.; Kawamoto, T.; Gehrmann, J.; Maguire, C.; et al. A Mouse Model of Congenital Heart Disease: Cardiac Arrhythmias and Atrial Septal Defect Caused by Haploinsufficiency of the Cardiac Transcription Factor Csx/Nkx2.5. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 317–326. [Google Scholar] [CrossRef]
- Terada, R.; Warren, S.; Lu, J.T.; Chien, K.R.; Wessels, A.; Kasahara, H. Ablation of Nkx2-5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc. Res. 2011, 91, 289–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, D.; Rasmussen, T.L.; Nakagawa, O.; McAnally, J.; Gottlieb, P.D.; Tucker, P.W.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development 2005, 132, 2669–2678. [Google Scholar] [CrossRef] [Green Version]
- Liberatore, C.M.; Searcy-Schrick, R.D.; Yutzey, K.E. Ventricular Expression of tbx5 Inhibits Normal Heart Chamber Development. Dev. Biol. 2000, 223, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshiba-Takeuchi, K.; Mori, A.D.; Kaynak, B.L.; Cebra-Thomas, J.; Sukonnik, T.; Georges, R.O.; Latham, S.; Beck, L.; Henkelman, R.M.; Black, B.L.; et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 2009, 461, 95–98. [Google Scholar] [CrossRef]
- Zhang, K.K.; Xiang, M.; Zhou, L.; Liu, J.; Curry, N.; Heine, S.D.; Garcia-Pavia, P.; Zhang, X.; Wang, Q.; Xie, L. Gene network and familial analyses uncover a gene network involving Tbx5/Osr1/Pcsk6 interaction in the second heart field for atrial septation. Hum. Mol. Genet. 2016, 25, 1140–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadeaua, M.; Georges, R.O.; Laforest, B.; Yamak, A.; Lefebvre, C.; Beauregard, J.; Paradis, P.; Bruneau, B.G.; Andelfinger, G.; Nemer, M. An endocardial pathway involving Tbx5, Gata4, and Nos3 required for atrial septum formation. Proc. Natl. Acad. Sci. USA 2010, 107, 19356–19361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, L.; Hoffmann, A.D.; Burnicka-Turek, O.; Friedland-Little, J.M.; Zhang, K.; Moskowitz, I.P. Tbx5-Hedgehog Molecular Networks Are Essential in the Second Heart Field for Atrial Septation. Dev. Cell 2012, 23, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Misra, C.; Chang, S.-W.; Basu, M.; Huang, N.; Garg, V. Disruption of myocardial Gata4 and Tbx5 results in defects in cardiomyocyte proliferation and atrioventricular septation. Hum. Mol. Genet. 2014, 23, 5025–5035. [Google Scholar] [CrossRef]
- Campione, M.; Steinbeisser, H.; Schweickert, A.; Deissler, K.; Van Bebber, F.; A Lowe, L.; Nowotschin, S.; Viebahn, C.; Haffter, P.; Kuehn, M.R.; et al. The homeobox gene Pitx2: Mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development 1999, 126, 1225–1234. [Google Scholar]
- Piedra, M.; Icardo, J.M.; Albajar, M.; Rodriguez-Rey, J.C.; A Ros, M. Pitx2 Participates in the Late Phase of the Pathway Controlling Left-Right Asymmetry. Cell 1998, 94, 319–324. [Google Scholar] [CrossRef] [Green Version]
- Ryan, A.K.; Blumberg, B.; Rodriguez-Esteban, C.; Yonei-Tamura, S.; Tamura, K.; Tsukui, T.; De La Peña, J.; Sabbagh, W.; Greenwald, J.; Choe, S.; et al. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Nat. Cell Biol. 1998, 394, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Logan, M.; Pagán-Westphal, S.M.; Smith, D.M.; Paganessi, L.; Tabin, C.J. The Transcription Factor Pitx2 Mediates Situs-Specific Morphogenesis in Response to Left-Right Asymmetric Signals. Cell 1998, 94, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, H.; Meno, C.; Koshiba, K.; Sugihara, M.; Itoh, H.; Ishimaru, Y.; Inoue, T.; Ohuchi, H.; Semina, E.V.; Murray, J.C.; et al. Pitx2, a Bicoid-Type Homeobox Gene, Is Involved in a Lefty-Signaling Pathway in Determination of Left-Right Asymmetry. Cell 1998, 94, 299–305. [Google Scholar] [CrossRef] [Green Version]
- Bisgrove, B.W.; Essner, J.J.; Yost, H.J. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development 2000, 127, 3567–3579. [Google Scholar] [PubMed]
- Collins, M.M.; Maischein, H.-M.; Dufourcq, P.; Charpentier, M.; Blader, P.; Stainier, D.Y. Pitx2c orchestrates embryonic axis extension via mesendodermal cell migration. eLife 2018, 7, e34880. [Google Scholar] [CrossRef]
- Dagle, J.; Sabel, J.L.; Littig, J.L.; Sutherland, L.B.; Kolker, S.J.; Weeks, D.L. Pitx2c attenuation results in cardiac defects and abnormalities of intestinal orientation in developing Xenopus laevis. Dev. Biol. 2003, 262, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Essner, J.J.; Branford, W.W.; Zhang, J.; Yost, H.J. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 2000, 127, 1081–1093. [Google Scholar]
- Gage, P.J.; Suh, H.; Camper, S.A. Dosage requirement of Pitx2 for development of multiple organs. Development 1999, 126, 4643–4651. [Google Scholar]
- Yu, X.; Amand, T.R.S.; Wang, S.; Li, G.; Zhang, Y.; Hu, Y.P.; Nguyen, L.; Qiu, M.S.; Chen, Y.P. Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development 2001, 128, 1005–1013. [Google Scholar] [PubMed]
- Lu, M.-F.; Pressman, C.L.; Dyer, R.; Johnson, R.L.; Martin, J.F. Function of Rieger syndrome gene in left–right asymmetry and craniofacial development. Nat. Cell Biol. 1999, 401, 276–278. [Google Scholar] [CrossRef]
- Kitamura, K.; Miura, H.; Miyagawa-Tomita, S.; Yanazawa, M.; Katoh-Fukui, Y.; Suzuki, R.; Ohuchi, H.; Suehiro, A.; Motegi, Y.; Nakahara, Y.; et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 1999, 126, 5749–5758. [Google Scholar]
- Shiratori, H.; Yashiro, K.; Shen, M.M.; Hamada, H. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development 2006, 133, 3015–3025. [Google Scholar] [CrossRef] [Green Version]
- A Lowe, L.; Yamada, S.; Kuehn, M.R. Genetic dissection of nodal function in patterning the mouse embryo. Development 2001, 128, 1831–1843. [Google Scholar]
- St Amand, T.R.; Ra, J.; Zhang, Y.; Hu, Y.; Baber, S.I.; Qiu, M.; Chen, Y. Cloning and expression pattern of chicken Pitx2: A new component in the SHH signaling pathway controlling embryonic heart looping. Biochem. Biophys. Res. Commun. 1998, 247, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Campione, M.; A Ros, M.; Icardo, J.M.; Piedra, E.; Christoffels, V.M.; Schweickert, A.; Blum, M.; Franco, D.; Moorman, A.F. Pitx2 Expression Defines a Left Cardiac Lineage of Cells: Evidence for Atrial and Ventricular Molecular Isomerism in the iv/iv Mice. Dev. Biol. 2001, 231, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Shiratori, H.; Sakuma, R.; Watanabe, M.; Hashiguchi, H.; Mochida, K.; Sakai, Y.; Nishino, J.; Saijoh, Y.; Whitman, M.; Hamada, H. Two-step regulation of left-right asymmetric expression of Pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol. Cell. 2001, 7, 137–149. [Google Scholar] [CrossRef]
- Campione, M.; Acosta, L.; Martinez, S.; Icardo, J.; Aranega, A.; Franco, D. Pitx2 and Cardiac Development: A Molecular Link between Left/Right Signaling and Congenital Heart Disease. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 89–95. [Google Scholar] [CrossRef]
- Franco, D.; Campione, M. The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc. Med. 2003, 13, 157–163. [Google Scholar] [CrossRef]
- Campione, M.; Franco, D. Current Perspectives in Cardiac Laterality. J. Cardiovasc. Dev. Dis. 2016, 3, 34. [Google Scholar] [CrossRef] [Green Version]
- Franco, D.; Sedmera, D.; Lozano-Velasco, E. Multiple Roles of Pitx2 in Cardiac Development and Disease. J. Cardiovasc. Dev. Dis. 2017, 4, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, D.; Liu, W.; Ma, L.; Dong, F.; Lu, M.-F.; Wang, D.; Verzi, M.P.; Cai, C.; Gage, P.J.; Evans, S.; et al. Pitx2 regulates cardiac left–right asymmetry by patterning second cardiac lineage-derived myocardium. Dev. Biol. 2006, 296, 437–449. [Google Scholar] [CrossRef] [Green Version]
- Franco, D.; Campione, M.; Kelly, R.G.; Zammit, P.S.; Buckingham, M.; Lamers, W.H.; Moorman, A.F.M. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ. Res. 2000, 87, 984–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mommersteeg, M.T.M.; Hoogaars, W.M.H.; Prall, O.W.J.; Vries, C.D.G.-D.; Wiese, C.; Clout, D.E.W.; Papaioannou, V.E.; Brown, N.A.; Harvey, R.P.; Moorman, A.F.M.; et al. Molecular Pathway for the Localized Formation of the Sinoatrial Node. Circ. Res. 2007, 100, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Ocaña, O.H.; Coskun, H.; Minguillón, C.; Murawala, P.; Tanaka, E.M.; Galcerán, J.; Muñoz-Chápuli, R.; Nieto, M.A. A right-handed signalling pathway drives heart looping in vertebrates. Nature 2017, 549, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Zeng, B.; Ren, X.-F.; Cao, F.; Zhou, X.-Y.; Zhang, J. Developmental patterns and characteristics of epicardial cell markers Tbx18 and Wt1 in murine embryonic heart. J. Biomed. Sci. 2011, 18, 67. [Google Scholar] [CrossRef] [Green Version]
- Moore, A.W.; McInnes, L.; Kreidberg, J.; Hastie, N.D.; Schedl, A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999, 126, 1845–1857. [Google Scholar]
- Acharya, A.; Baek, S.T.; Huang, G.; Eskiocak, B.; Goetsch, S.; Sung, C.Y.; Banfi, S.; Sauer, M.F.; Olsen, G.S.; Duffield, J.S.; et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012, 139, 2139–2149. [Google Scholar] [CrossRef] [Green Version]
- Ishii, Y.; Langberg, J.D.; Hurtado, R.; Lee, S.; Mikawa, T. Induction of proepicardial marker gene expression by the liver bud. Development 2007, 134, 3627–3637. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Hoogaars, W.M.; Barnett, P.; Grieskamp, T.; Rana, M.S.; Buermans, H.; Farin, H.F.; Petry, M.; Heallen, T.; Martin, J.F.; et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol. Life Sci. 2012, 69, 1377–1389. [Google Scholar] [CrossRef] [Green Version]
- Christoffels, V.M.; Hoogaars, W.M.; Tessari, A.; Clout, D.E.; Moorman, A.F.M.; Campione, M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn. 2004, 229, 763–770. [Google Scholar] [CrossRef]
- Sedletcaia, A.; Evans, T. Heart chamber size in zebrafish is regulated redundantly by duplicated tbx2 genes. Dev. Dyn. 2011, 240, 1548–1557. [Google Scholar] [CrossRef] [Green Version]
- Chi, N.C.; Shaw, R.M.; De Val, S.; Kang, G.; Jan, L.Y.; Black, B.L.; Stainier, D.Y. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008, 22, 734–739. [Google Scholar] [CrossRef] [Green Version]
- Shirai, M.; Imanaka-Yoshida, K.; Schneider, M.D.; Schwartz, R.J.; Morisaki, T. T-box 2, a mediator of Bmp-Smad signaling, induced hyaluronan synthase 2 and Tgfbeta2 expression and endocardial cushion formation. Proc. Natl. Acad. Sci. USA 2009, 106, 18604–18609. [Google Scholar] [CrossRef] [Green Version]
- Dupays, L.; Kotecha, S.; Angst, B.; Mohun, T.J. Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev. Biol. 2009, 333, 121–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoogaars, W.M.; Tessari, A.; Moorman, A.F.; de Boer, P.A.; Hagoort, J.; Soufan, A.T.; Campione, M.; Christoffels, V.M. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004, 62, 489–499. [Google Scholar] [CrossRef]
- Mesbah, K.; Harrelson, Z.; Théveniau-Ruissy, M.; Papaioannou, V.E.; Kelly, R.G. Tbx3 Is Required for Outflow Tract Development. Circ. Res. 2008, 103, 743–750. [Google Scholar] [CrossRef]
- Mohan, R.A.; Mommersteeg, M.T.M.; Domínguez, J.N.; Choquet, C.; Wakker, V.; de Gier-de Vries, C.; Boink, G.J.J.; Boukens, B.J.; Miquerol, L.; Verkerk, A.O.; et al. Embryonic Tbx3+ cardiomyocytes form the mature cardiac conduction system by progressive fate restriction. Development 2018, 145, dev167361. [Google Scholar] [CrossRef] [Green Version]
- Mohan, R.A.; Bosada, F.M.; Van Weerd, J.H.; Van Duijvenboden, K.; Wang, J.; Mommersteeg, M.T.; Hooijkaas, I.B.; Wakker, V.; Vries, C.D.G.-D.; Coronel, R.; et al. T-box transcription factor 3 governs a transcriptional program for the function of the mouse atrioventricular conduction system. Proc. Natl. Acad. Sci. USA 2020, 117, 18617–18626. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Nomura-Kitabayashi, A.; Cai, W.; Yan, J.; Christoffels, V.M.; Cai, C.-L. Myocardial Tbx20 regulates early atrioventricular canal formation and endocardial epithelial–mesenchymal transition via Bmp2. Dev. Biol. 2011, 360, 381–390. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.K.; Christoffels, V.M.; Dias, J.M.; Trowe, M.-O.; Petry, M.; Schuster-Gossler, K.; Bürger, A.; Ericson, J.; Kispert, A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development 2005, 132, 2697–2707. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Horsthuis, T.; Farin, H.F.; Grieskamp, T.; Norden, J.; Petry, M.; Wakker, V.; Moorman, A.F.M.; Christoffels, V.M.; Kispert, A. Tbx20 Interacts With Smads to Confine Tbx2 Expression to the Atrioventricular Canal. Circ. Res. 2009, 105, 442–452. [Google Scholar] [CrossRef] [Green Version]
- Stennard, F.A.; Costa, M.W.; Lai, D.; Biben, C.; Furtado, M.B.; Solloway, M.J.; McCulley, D.J.; Leimena, C.; Preis, J.I.; Dunwoodie, S.L.; et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 2005, 132, 2451–2462. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Yutzey, K.E. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev. Biol. 2012, 363, 234–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greulich, F.; Rudat, C.; Kispert, A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011, 91, 212–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plageman, T.F.; Yutzey, K.E. T-box genes and heart development: Putting the “T” in heart. Dev. Dyn. 2005, 232, 11–20. [Google Scholar] [CrossRef]
- Cai, C.-L.; Zhou, W.; Yang, L.; Bu, L.; Qyang, Y.; Zhang, X.; Li, X.; Rosenfeld, M.G.; Chen, J.; Evans, S.M. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 2005, 132, 2475–2487. [Google Scholar] [CrossRef] [Green Version]
- Christoffels, V.M.; Keijser, A.G.; Houweling, A.C.; Clout, D.E.; Moorman, A.F. Patterning the Embryonic Heart: Identification of Five Mouse Iroquois Homeobox Genes in the Developing Heart. Dev. Biol. 2000, 224, 263–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.-H.; Rosen, A.; Bruneau, B.G.; Hui, C.-C.; Backx, P.H. Iroquois Homeodomain Transcription Factors in Heart Development and Function. Circ. Res. 2012, 110, 1513–1524. [Google Scholar] [CrossRef] [Green Version]
- Bruneau, B.G.; Bao, Z.-Z.; Tanaka, M.; Schott, J.-J.; Izumo, S.; Cepko, C.L.; Seidman, J.; Seidman, C.E. Cardiac Expression of the Ventricle-Specific Homeobox Gene Irx4 Is Modulated by Nkx2-5 and dHand. Dev. Biol. 2000, 217, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Kokubo, H.; Miyagawa-Tomita, S.; Nakazawa, M.; Saga, Y.; Johnson, R.L. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev. Biol. 2005, 278, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Kokubo, H.; Tomita-Miyagawa, S.; Hamada, Y.; Saga, Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development 2007, 134, 747–755. [Google Scholar] [CrossRef] [Green Version]
- Rutenberg, J.B.; Fischer, A.; Jia, H.; Gessler, M.; Zhong, T.P.; Mercola, M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 2006, 133, 4381–4390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koibuchi, N.; Chin, M.T. CHF1/Hey2 Plays a Pivotal Role in Left Ventricular Maturation Through Suppression of Ectopic Atrial Gene Expression. Circ. Res. 2007, 100, 850–855. [Google Scholar] [CrossRef] [Green Version]
- Pereira, F.A.; Qiu, Y.; Zhou, G.; Tsai, M.-J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999, 13, 1037–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, D.; Cserjesi, P.; Olson, E.N. A Subclass of bHLH Proteins Required for Cardiac Morphogenesis. Science 1995, 270, 1995–1999. [Google Scholar] [CrossRef]
- Srivastava, D.; Thomas, T.; Lin, Q.; Kirby, M.L.; Brown, R.; Olson, E.N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997, 16, 154–160. [Google Scholar] [CrossRef]
- Togi, K.; Kawamoto, T.; Yamauchi, R.; Yoshida, Y.; Kita, T.; Tanaka, M. Role of Hand1/eHAND in the Dorso-Ventral Patterning and Interventricular Septum Formation in the Embryonic Heart. Mol. Cell. Biol. 2004, 24, 4627–4635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagishi, H.; Yamagishi, C.; Nakagawa, O.; Harvey, R.P.; Olson, E.N.; Srivastava, D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev. Biol. 2001, 239, 190–203. [Google Scholar] [CrossRef] [Green Version]
- Bolte, C.; Zhang, Y.; Wang, I.-C.; Kalin, T.V.; Molkentin, J.D.; Kalinichenko, V.V. Expression of Foxm1 Transcription Factor in Cardiomyocytes Is Required for Myocardial Development. PLoS ONE 2011, 6, e22217. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Kook, H.; Milewski, R.; Gitler, A.D.; Lu, M.M.; Li, J.; Nazarian, R.; Schnepp, R.; Jen, K.; Biben, C.; et al. Hop Is an Unusual Homeobox Gene that Modulates Cardiac Development. Cell 2002, 110, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Lavallée, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; E Horb, M.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213. [Google Scholar] [CrossRef]
- Niu, Z.; Yu, W.; Zhang, S.X.; Barron, M.; Belaguli, N.S.; Schneider, M.D.; Parmacek, M.; Nordheim, A.; Schwartz, R.J. Conditional Mutagenesis of the Murine Serum Response Factor Gene Blocks Cardiogenesis and the Transcription of Downstream Gene Targets. J. Biol. Chem. 2005, 280, 32531–32538. [Google Scholar] [CrossRef] [Green Version]
- Parlakian, A.; Tuil, D.; Hamard, G.; Tavernier, G.; Hentzen, D.; Concordet, J.-P.; Paulin, D.; Li, Z.; Daegelen, D. Targeted Inactivation of Serum Response Factor in the Developing Heart Results in Myocardial Defects and Embryonic Lethality. Mol. Cell. Biol. 2004, 24, 5281–5289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza-Lewis, R.A.; Yu, L.; He, F.; Liu, H.; Tang, R.; Shi, J.; Sun, X.; Martin, J.F.; Wang, D.; Yang, J.; et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009, 327, 376–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.; Xin, Y.; Zhao, Y.; Hu, J. Shox2: The Role in Differentiation and Development of Cardiac Conduction System. Tohoku J. Exp. Med. 2018, 244, 177–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, W.; Wang, J.; Song, Y.; Yu, D.; Sun, C.; Liu, C.; Chen, F.; Zhang, Y.; Wang, F.; Harvey, R.P.; et al. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 2015, 142, 2521–2532. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Espinoza-Lewis, R.A.; Chen, C.; Hu, X.; Zhang, Y.; Chen, Y.-P. The Role of Shox2 in SAN Development and Function. Pediatr. Cardiol. 2012, 33, 882–889. [Google Scholar] [CrossRef]
- Liu, H.; Chen, C.-H.; Espinoza-Lewis, R.A.; Jiao, Z.; Sheu, I.; Hu, X.; Lin, M.; Zhang, Y.; Chen, Y.-P. Functional Redundancy between HumanSHOXand MouseShox2Genes in the Regulation of Sinoatrial Node Formation and Pacemaking Function. J. Biol. Chem. 2011, 286, 17029–17038. [Google Scholar] [CrossRef] [Green Version]
- Blaschke, R.J.; Hahurij, N.D.; Kuijper, M.S.; Just, S.; Wisse, L.J.; Deissler, K.; Maxelon, T.; Anastassiadis, K.; Spitzer, J.; Hardt, S.E.; et al. Targeted Mutation Reveals Essential Functions of the Homeodomain Transcription Factor Shox2 in Sinoatrial and Pacemaking Development. Circulation 2007, 115, 1830–1838. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Lan, Y.; Cho, E.-S.; Maltby, K.M.; Jiang, R. Odd-skipped related 1 (Odd1) is an essential regulator of heart and urogenital development. Dev. Biol. 2005, 288, 582–594. [Google Scholar] [CrossRef] [Green Version]
- Chiplunkar, A.R.; Lung, T.K.; Alhashem, Y.; Koppenhaver, B.A.; Salloum, F.N.; Kukreja, R.C.; Haar, J.L.; Lloyd, J.A. Krüppel-Like Factor 2 Is Required for Normal Mouse Cardiac Development. PLoS ONE 2013, 8, e54891. [Google Scholar] [CrossRef] [Green Version]
- Gawdzik, J.C.; Yue, M.S.; Martin, N.R.; Elemans, L.M.; Lanham, K.A.; Heideman, W.; Rezendes, R.; Baker, T.R.; Taylor, M.R.; Plavicki, J.S. sox9b is required in cardiomyocytes for cardiac morphogenesis and function. Sci. Rep. 2018, 8, 13906. [Google Scholar] [CrossRef]
- Moskowitz, I.P.; Wang, J.; Peterson, M.A.; Pu, W.T.; MacKinnon, A.C.; Oxburgh, L.; Chu, G.C.; Sarkar, M.; Berul, C.; Smoot, L.; et al. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. Proc. Natl. Acad. Sci. USA 2011, 108, 4006–4011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, E.A.; Vitelli, F.; Su, H.; Morishima, M.; Huynh, T.; Pramparo, T.; Jurecic, V.; Ogunrinu, G.; Sutherland, H.F.; Scambler, P.J.; et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 2001, 410, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Kume, T.; Jiang, H.; Topczewska, J.M.; Hogan, B.L. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001, 15, 2470–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergwerff, M.; Groot, A.G.-D.; Wisse, L.J.; DeRuiter, M.C.; Wessels, A.; Martin, J.F.; Olson, E.N.; Kern, M.J. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000, 436, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Xu, J.; Molkentin, J.D. Re-employment of developmental transcription factors in adult heart disease. Semin. Cell Dev. Biol. 2007, 18, 117–131. [Google Scholar] [CrossRef] [Green Version]
- Hamada, H. Molecular and cellular basis of left-right asymmetry in vertebrates. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 273–296. [Google Scholar] [CrossRef]
- Chazaud, C.; Chambon, P.; Dollé, P. Retinoic acid is required in the mouse embryo for left-right asymmetry determination and heart morphogenesis. Development 1999, 126, 2589–2596. [Google Scholar]
- Zile, M.H.; Kostetskii, I.; Yuan, S.; Kostetskaia, E.; Amand, T.R.S.; Chen, Y.; Jiang, W. Retinoid Signaling Is Required to Complete the Vertebrate Cardiac Left/Right Asymmetry Pathway. Dev. Biol. 2000, 223, 323–338. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, H.; Miyakawa, N.; Miyake, A.; Itoh, N. Fgf4 is required for left–right patterning of visceral organs in zebrafish. Dev. Biol. 2009, 332, 177–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyers, E.N.; Martin, G.R. Differences in left-right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science 1999, 285, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Viebahn, C.; Blum, M. FGF8 Acts as a Right Determinant during Establishment of the Left-Right Axis in the Rabbit. Curr. Biol. 2002, 12, 1807–1816. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, H.; Rebagliati, M.; Ahmad, N.; Muraoka, O.; Kurokawa, T.; Hibi, M.; Suzuki, T. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 2004, 131, 1741–1753. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, R.; Van Dinther, M.; Bakkers, J.; Wilkinson, R.N.; Patient, R.; Dijke, P.T.; Mummery, C. Two novel type II receptors mediate BMP signalling and are required to establish left–right asymmetry in zebrafish. Dev. Biol. 2008, 315, 55–71. [Google Scholar] [CrossRef] [Green Version]
- Meno, C.; Shimono, A.; Saijoh, Y.; Yashiro, K.; Mochida, K.; Ohishi, S.; Noji, S.; Kondoh, H.; Hamada, H. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 1998, 94, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Mironova, E.; Whitaker, L.L.; Edwards, L.; Yost, H.J.; Ramsdell, A.F. ALK4 functions as a receptor for multiple TGF beta-related ligands to regulate left-right axis determination and mesoderm induction in Xenopus. Dev. Biol. 2004, 268, 280–294. [Google Scholar] [CrossRef] [Green Version]
- Branford, W.W.; Essner, J.J.; Yost, H. Regulation of Gut and Heart Left–Right Asymmetry by Context-Dependent Interactions between Xenopus Lefty and BMP4 Signaling. Dev. Biol. 2000, 223, 291–306. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Zwijsen, A.; Vogel, H.; Huylebroeck, D.; Matzuk, M.M.; Vogel, H. Smad5 Is Essential for Left–Right Asymmetry in Mice. Dev. Biol. 2000, 219, 71–78. [Google Scholar] [CrossRef]
- Tsiairis, C.; McMahon, A.P. An Hh-Dependent Pathway in Lateral Plate Mesoderm Enables the Generation of Left/Right Asymmetry. Curr. Biol. 2009, 19, 1912–1917. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Qi, J. miR-430a regulates the development of left–right asymmetry by targeting sqt in the teleost. Gene 2020, 745, 144628. [Google Scholar] [CrossRef] [PubMed]
- Heigwer, J.; Kutzner, J.; Haeussler, M.; Burkhalter, M.D.; Draebing, T.; Juergensen, L.; Katus, H.A.; Philipp, M.; Westhoff, J.H.; Hassel, D. miR-103/107 regulates left-right asymmetry in zebrafish by modulating Kupffer’s vesicle development and ciliogenesis. Biochem. Biophys. Res. Commun. 2020, 527, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Hatayama, M.; Mikoshiba, K.; Aruga, J. IP3 signaling is required for cilia formation and left–right body axis determination in Xenopus embryos. Biochem. Biophys. Res. Commun. 2011, 410, 520–524. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Tsai, M.-Y.; Liu, Y.-H.; Lu, Y.-F.; Chen, Y.-C.; Lai, Y.-R.; Liao, H.-C.; Lien, H.-W.; Yang, C.-H.; Huang, C.-J.; et al. Klf8 regulates left-right asymmetric patterning through modulation of Kupffer’s vesicle morphogenesis and spaw expression. J. Biomed. Sci. 2017, 24, 45. [Google Scholar] [CrossRef] [PubMed]
- Rankin, C.T.; Bunton, T.; Lawler, A.M.; Lee, S.-J. Regulation of left-right patterning in mice by growth/differentiation factor-1. Nat. Genet. 2000, 24, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Tamakoshi, T.; Itakura, T.; Chandra, A.; Uezato, T.; Yang, Z.; Xue, X.-D.; Wang, B.; Hackett, B.P.; Yokoyama, T.; Miura, N. Roles of the Foxj1 and Inv genes in the left–right determination of internal organs in mice. Biochem. Biophys. Res. Commun. 2006, 339, 932–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gormley, J.P.; Nascone-Yoder, N.M. Left and right contributions to the Xenopus heart: Implications for asymmetric morphogenesis. Dev. Genes Evol. 2003, 213, 390–398. [Google Scholar] [CrossRef]
- Walton, R.Z.; Bruce, A.E.; Olivey, H.E.; Najib, K.; Johnson, V.; Earley, J.U.; Ho, R.K.; Svensson, E.C. Fog1 is required for cardiac looping in zebrafish. Dev. Biol. 2006, 289, 482–493. [Google Scholar] [CrossRef] [Green Version]
- Duboc, V.; Röttinger, E.; Lapraz, F.; Besnardeau, L.; Lepage, T. Left-Right Asymmetry in the Sea Urchin Embryo Is Regulated by Nodal Signaling on the Right Side. Dev. Cell 2005, 9, 147–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, S.; Ahmad, N.; Rebagliati, M. Zebrafish Hearts and Minds: Nodal Signaling in Cardiac and Neural Left-Right Asymmetry. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 27–36. [Google Scholar] [CrossRef]
- Chin, A.J.; Tsang, M.; Weinberg, E.S. Heart and Gut Chiralities Are Controlled Independently from Initial Heart Position in the Developing Zebrafish. Dev. Biol. 2000, 227, 403–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, A.; Mijalski, T.; Schlange, T.; Dai, W.; Overbeek, P.A.; Arnold, H.-H.; Brand, T. The homeobox gene it NKX3.2 is a target of left–right signalling and is expressed on opposite sides in chick and mouse embryos. Curr. Biol. 1999, 9, 911–914. [Google Scholar] [CrossRef] [Green Version]
- Soukup, V. Left-right asymmetry specification in amphioxus: Review and prospects. Int. J. Dev. Biol. 2017, 61, 611–620. [Google Scholar] [CrossRef]
- Tadjuidje, E.; Kofron, M.; Mir, A.; Wylie, C.; Heasman, J.; Cha, S.-W. Nodal signalling in Xenopus: The role of Xnr5 in left/right asymmetry and heart development. Open Biol. 2016, 6, 150187. [Google Scholar] [CrossRef] [Green Version]
- Hamada, H.; Meno, C.; Saijoh, Y.; Adachi, H.; Yashiro, K.; Sakuma, R.; Shiratori, H. Role of asymmetric signals in left-right patterning in the mouse. Am. J. Med. Genet. 2001, 101, 324–327. [Google Scholar] [CrossRef]
- Boorman, C.J.; Shimeld, S.M. The evolution of left-right asymmetry in chordates. BioEssays 2002, 24, 1004–1011. [Google Scholar] [CrossRef]
- Grimes, D.T.; Burdine, R.D. Left–Right Patterning: Breaking Symmetry to Asymmetric Morphogenesis. Trends Genet. 2017, 33, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Rago, L.; Castroviejo, N.; Fazilaty, H.; Garcia-Asencio, F.; Ocaña, O.H.; Galcerán, J.; Nieto, M.A. MicroRNAs Establish the Right-Handed Dominance of the Heart Laterality Pathway in Vertebrates. Dev. Cell 2019, 51, 446–459.e5. [Google Scholar] [CrossRef]
- Dasgupta, A.; Amack, J.D. Cilia in vertebrate left–right patterning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150410. [Google Scholar] [CrossRef] [Green Version]
- Blum, M.; Vick, P. Left–Right Asymmetry: Cilia and Calcium Revisited. Curr. Biol. 2015, 25, R205–R207. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.R.; Pakula, G.; Klaeyle, L.; Fukui, H.; Vilfan, A.; Supatto, W.; Vermot, J. Chiral Cilia Orientation in the Left-Right Organizer. Cell Rep. 2018, 25, 2008–2016.e4. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, K.; Hamada, H. Cilia in Left–Right Symmetry Breaking. Cold Spring Harb. Perspect. Biol. 2017, 9, a028282. [Google Scholar] [CrossRef]
- Chien, Y.-H.; Srinivasan, S.; Keller, R.; Kintner, C. Mechanical Strain Determines Cilia Length, Motility, and Planar Position in the Left-Right Organizer. Dev. Cell 2018, 45, 316–330.e4. [Google Scholar] [CrossRef] [Green Version]
- Yoshiba, S.; Hamada, H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left–right symmetry. Trends Genet. 2014, 30, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, A.; Miyamoto, T.; Simono, F.; Kurogi, N.; Shirae-Kurabayashi, M.; Awazu, A.; Suzuki, K.-I.T.; Yamamoto, T.; Sakamoto, N. Cilia play a role in breaking left-right symmetry of the sea urchin embryo. Genes Cells 2016, 21, 568–578. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Shi, C.; Zhong, Y.; Liu, X.; Yan, Q.; Wu, X.; Wang, Y.; Li, G. Cilia-driven asymmetric Hedgehog signalling determines the amphioxus left-right axis by controlling Dand5 expression. Development 2020, 147, dev182469. [Google Scholar] [CrossRef]
- Yuan, S.; Brueckner, M. Visualization and Manipulation of Cilia and Intraciliary Calcium in the Zebrafish Left–Right Organizer. Methods Mol Biol. 2016, 1454, 123–147. [Google Scholar] [CrossRef]
- Pereira, R.; Barbosa, T.; Gales, L.; Oliveira, E.; Santos, R.; Oliveira, J.; Sousa, M. Clinical and Genetic Analysis of Children with Kartagener Syndrome. Cells 2019, 8, 900. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Huang, Q.; Zhu, P.; Zhao, P.; Tan, X.; Liu, S.; Li, S.; Han, X.; Cheng, L.; Li, B.; et al. Identification of Pathogenic Mutations and Investigation of the NOTCH Pathway Activation in Kartagener Syndrome. Front. Genet. 2019, 10, 749. [Google Scholar] [CrossRef] [Green Version]
- Bellchambers, H.M.; Ware, S.M. ZIC3 in Heterotaxy. Adv. Exp. Med. Biol. 2018, 1046, 301–327. [Google Scholar] [CrossRef]
- Horváth, J.; Fliegauf, M.; Olbrich, H.; Kispert, A.; King, S.M.; Mitchison, H.; Zariwala, M.A.; Knowles, M.R.; Sudbrak, R.; Fekete, G.; et al. Identification and Analysis of Axonemal Dynein Light Chain 1 in Primary Ciliary Dyskinesia Patients. Am. J. Respir. Cell Mol. Biol. 2005, 33, 41–47. [Google Scholar] [CrossRef]
- Mazor, M.; Alkrinawi, S.; Chalifa-Caspi, V.; Manor, E.; Sheffield, V.C.; Aviram, M.; Parvari, R. Primary Ciliary Dyskinesia Caused by Homozygous Mutation in DNAL1, Encoding Dynein Light Chain 1. Am. J. Hum. Genet. 2011, 88, 599–607. [Google Scholar] [CrossRef] [Green Version]
- Loges, N.T.; Olbrich, H.; Fenske, L.; Mussaffi, H.; Horvath, J.; Fliegauf, M.; Kuhl, H.; Baktai, G.; Peterffy, E.; Chodhari, R.; et al. DNAI2 Mutations Cause Primary Ciliary Dyskinesia with Defects in the Outer Dynein Arm. Am. J. Hum. Genet. 2008, 83, 547–558. [Google Scholar] [CrossRef] [Green Version]
- Olbrich, H.; Häffner, K.; Kispert, A.; Völkel, A.; Volz, A.; Sasmaz, G.; Reinhardt, R.; Hennig, S.; Lehrach, H.; Konietzko, N.; et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat. Genet. 2002, 30, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, G.W.; Loges, N.T.; Klinkenbusch, J.A.; Olbrich, H.; Pennekamp, P.; Menchen, T.; Raidt, J.; Wallmeier, J.; Werner, C.; Westermann, C.; et al. DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes. Am. J. Respir. Cell Mol. Biol. 2016, 55, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yagi, H.; Onuoha, E.O.; Damerla, R.R.; Francis, R.; Furutani, Y.; Tariq, M.; King, S.M.; Hendricks, G.; Cui, C.; et al. DNAH6 and Its Interactions with PCD Genes in Heterotaxy and Primary Ciliary Dyskinesia. PLoS Genet. 2016, 12, e1005821. [Google Scholar] [CrossRef] [Green Version]
- Loges, N.T.; Olbrich, H.; Becker-Heck, A.; Häffner, K.; Heer, A.; Reinhard, C.; Schmidts, M.; Kispert, A.; Zariwala, M.A.; Leigh, M.W.; et al. Deletions and Point Mutations of LRRC50 Cause Primary Ciliary Dyskinesia Due to Dynein Arm Defects. Am. J. Hum. Genet. 2009, 85, 883–889. [Google Scholar] [CrossRef] [Green Version]
- Merveille, A.C.; Davis, E.E.; Becker-Heck, A.; Legendre, M.; Amirav, I.; Bataille, G.; Belmont, J.; Beydon, N.; Billen, F.; Clément, A.; et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2011, 43, 72–78. [Google Scholar] [CrossRef]
- Wallmeier, J.; Shiratori, H.; Dougherty, G.W.; Edelbusch, C.; Hjeij, R.; Loges, N.T.; Menchen, T.; Olbrich, H.; Pennekamp, P.; Raidt, J.; et al. TTC25 Deficiency Results in Defects of the Outer Dynein Arm Docking Machinery and Primary Ciliary Dyskinesia with Left-Right Body Asymmetry Randomization. Am. J. Hum. Genet. 2016, 99, 460–469. [Google Scholar] [CrossRef]
- Hirokawa, N.; Tanaka, Y.; Okada, Y. Left-Right Determination: Involvement of Molecular Motor KIF3, Cilia, and Nodal Flow. Cold Spring Harb. Perspect. Biol. 2009, 1, a000802. [Google Scholar] [CrossRef] [Green Version]
- Jahr, M.; Schlueter, J.; Brand, T.; Männer, J. Development of the proepicardium in Xenopus laevis. Dev. Dyn. 2008, 237, 3088–3096. [Google Scholar] [CrossRef]
- Niderla-Bielińska, J.; Jankowska-Steifer, E.; Flaht-Zabost, A.; Gula, G.; Czarnowska, E.; Ratajska, A. Proepicardium: Current Understanding of its Structure, Induction, and Fate. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2019, 302, 893–903. [Google Scholar] [CrossRef]
- Maya-Ramos, L.; Cleland, J.; Bressan, M.; Mikawa, T. Induction of the Proepicardium. J. Dev. Biol. 2013, 1, 82–91. [Google Scholar] [CrossRef] [Green Version]
- Niderla-Bielińska, J.; Ciszek, B.; Jankowska-Steifer, E.; Flaht-Zabost, A.; Gula, G.; Radomska-Leśniewska, D.M.; Ratajska, A. Mouse Proepicardium Exhibits a Sprouting Response to Exogenous Proangiogenic Growth Factors in vitro. J. Vasc. Res. 2016, 53, 83–93. [Google Scholar] [CrossRef]
- Rodgers, L.S.; Lalani, S.; Runyan, R.B.; Camenisch, T.D. Differential growth and multicellular villi direct proepicardial translocation to the developing mouse heart. Dev. Dyn. 2008, 237, 145–152. [Google Scholar] [CrossRef]
- Nahirney, P.C.; Mikawa, T.; Fischman, D.A. Evidence for an extracellular matrix bridge guiding proepicardial cell migration to the myocardium of chick embryos. Dev. Dyn. 2003, 227, 511–523. [Google Scholar] [CrossRef]
- Plavicki, J.S.; Hofsteen, P.; Yue, M.S.; A Lanham, K.; Peterson, R.E.; Heideman, W. Multiple modes of proepicardial cell migration require heartbeat. BMC Dev. Biol. 2014, 14, 18. [Google Scholar] [CrossRef] [Green Version]
- Schlueter, J.; Männer, J.; Brand, T. BMP is an important regulator of proepicardial identity in the chick embryo. Dev. Biol. 2006, 295, 546–558. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Stainier, D.Y. Tbx5 and Bmp Signaling Are Essential for Proepicardium Specification in Zebrafish. Circ. Res. 2010, 106, 1818–1828. [Google Scholar] [CrossRef] [Green Version]
- Ishii, Y.; Garriock, R.J.; Navetta, A.M.; Coughlin, L.E.; Mikawa, T. BMP Signals Promote Proepicardial Protrusion Necessary for Recruitment of Coronary Vessel and Epicardial Progenitors to the Heart. Dev. Cell 2010, 19, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Andrés-Delgado, L.; Ernst, A.; Galardi-Castilla, M.; Bazaga, D.; Peralta, M.; Münch, J.; González-Rosa, J.M.; Marques, I.; Tessadori, F.; De La Pompa, J.L.; et al. Actin dynamics and the Bmp pathway drive apical extrusion of proepicardial cells. Development 2019, 146, dev174961. [Google Scholar] [CrossRef] [Green Version]
- Andrés-Delgado, L.; Galardi-Castilla, M.; Münch, J.; Peralta, M.; Ernst, A.; González-Rosa, J.M.; Tessadori, F.; Santamaría, L.; Bakkers, J.; Vermot, J.; et al. Notch and Bmp signaling pathways act coordinately during the formation of the proepicardium. Dev. Dyn. 2020, 249, 1455–1469. [Google Scholar] [CrossRef]
- Schlueter, J.; Brand, T. A right-sided pathway involving FGF8/Snai1 controls asymmetric development of the proepicardium in the chick embryo. Proc. Natl. Acad. Sci. SUA 2009, 106, 7485–7490. [Google Scholar] [CrossRef] [Green Version]
- Torlopp, A.; Schlueter, J.; Brand, T. Role of fibroblast growth factor signaling during proepicardium formation in the chick embryo. Dev. Dyn. 2010, 239, 2393–2403. [Google Scholar] [CrossRef]
- Li, J.; Miao, L.; Zhao, C.; Shaikh, Q.W.M.; Shieh, D.; Guo, H.; Lu, Y.; Hu, S.; Huang, A.; Zhang, L.; et al. CDC42 is required for epicardial and pro-epicardial development by mediating FGF receptor trafficking to the plasma membrane. Development 2017, 144, 1635–1647. [Google Scholar] [CrossRef] [Green Version]
- Kruithof, B.P.; Van Wijk, B.; Somi, S.; Kruithof-de Julio, M.; Pérez Pomares, J.M.; Weesie, F.; Wessels, A.; Moorman, A.F.; van der Hoff, M.J. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev. Biol. 2006, 295, 507–522. [Google Scholar] [CrossRef] [Green Version]
- Dueñas, A.; Aranega, A.E.; Franco, D. More than Just a Simple Cardiac Envelope; Cellular Contributions of the Epicardium. Front. Cell Dev. Biol. 2017, 5, 44. [Google Scholar] [CrossRef] [Green Version]
- Carmona, R.; Guadix, J.A.; Cano, E.; Ruiz-Villalba, A.; Portillo-Sánchez, V.; Pérez-Pomares, J.M.; Pérez-Pomares, R. The embryonic epicardium: An essential element of cardiac development. J. Cell Mol. Med. 2010, 14, 2066–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peralta, M.; González-Rosa, J.M.; Marques, I.J.; Mercader, N. The Epicardium in the Embryonic and Adult Zebrafish. J. Dev. Biol. 2014, 2, 101–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Männer, J.; Pérez-Pomares, J.; Macías, D.; Muñoz-Chápuli, R. The Origin, Formation and Developmental Significance of the Epicardium: A Review. Cells Tissues Organs 2001, 169, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pomares, J.M.; de la Pompa, J.L. Signaling during epicardium and coronary vessel development. Circ. Res. 2011, 109, 1429–1442. [Google Scholar] [CrossRef] [Green Version]
- Olivey, H.E.; Compton, L.A.; Barnett, J.V. Coronary Vessel DevelopmentThe Epicardium Delivers. Trends Cardiovasc. Med. 2004, 14, 247–251. [Google Scholar] [CrossRef]
- Cano, E.; Carmona, R.; Ruiz-Villalba, A.; Rojas, A.; Chau, Y.-Y.; Wagner, K.D.; Wagner, N.; Hastie, N.D.; Muñoz-Chápuli, R.; Pérez-Pomares, J.M. Extracardiac septum transversum/proepicardial endothelial cells pattern embryonic coronary arterio-venous connections. Proc. Natl. Acad. Sci. USA 2016, 113, 656–661. [Google Scholar] [CrossRef] [Green Version]
- Carmona, R.; Barrena, S.; Gambero, A.J.L.; Rojas, A.; Muñoz-Chápuli, R. Epicardial cell lineages and the origin of the coronary endothelium. FASEB J. 2020, 34, 5223–5239. [Google Scholar] [CrossRef] [Green Version]
- Zamora, M.; Männer, J.; Ruiz-Lozano, P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc. Natl. Acad. Sci. USA 2007, 104, 18109–18114. [Google Scholar] [CrossRef] [Green Version]
- Smart, N.; Riley, P.R. The epicardium as a candidate for heart regeneration. Future Cardiol. 2012, 8, 53–69. [Google Scholar] [CrossRef] [Green Version]
- Kathiriya, I.S.; Srivastava, D. Left-right asymmetry and cardiac looping: Implications for cardiac development and congenital heart disease. Am. J. Med. Genet. 2000, 97, 271–279. [Google Scholar] [CrossRef]
- Anderson, R.H.; Brown, N.A.; Meno, C.; Spicer, D. The importance of being isomeric. Clin. Anat. 2015, 28, 477–486. [Google Scholar] [CrossRef]
- Bartram, U.; Wirbelauer, J.; Speer, C.P. Heterotaxy Syndrome—Asplenia and Polysplenia as Indicators of Visceral Malposition and Complex Congenital Heart Disease. Biol. Neonate 2005, 88, 278–290. [Google Scholar] [CrossRef]
- Belmont, J.W.; Mohapatra, B.; Towbin, J.A.; Ware, S.M. Molecular genetics of heterotaxy syndromes. Curr. Opin. Cardiol. 2004, 19, 216–220. [Google Scholar] [CrossRef]
- Degenhardt, K.; Rychik, J. Fetal Situs, Isomerism, Heterotaxy Syndrome: Diagnostic Evaluation and Implication for Postnatal Management. Curr. Treat Options Cardiovasc. Med. 2016, 18, 77. [Google Scholar] [CrossRef]
- Kothari, S.S. Non-cardiac issues in patients with heterotaxy syndrome. Ann. Pediatr. Cardiol. 2014, 7, 187–192. [Google Scholar] [CrossRef]
- Mishra, S. Cardiac and Non-Cardiac Abnormalities in Heterotaxy Syndrome. Indian J. Pediatr. 2015, 82, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Phoon, C.K.; Neill, C.A. Asplenia syndrome: Insight into embryology through an analysis of cardiac and extracardiac anomalies. Am. J. Cardiol. 1994, 73, 581–587. [Google Scholar] [CrossRef]
- Sutherland, M.J.; Ware, S.M. Disorders of left-right asymmetry: Heterotaxy and situs inversus. Am. J. Med. Genet. Part C Semin. Med. Genet. 2009, 15, 307–317. [Google Scholar] [CrossRef]
- Tawfik, A.; Batouty, N.M.; Zaky, M.M.; Eladalany, M.A.; Elmokadem, A.H. Polysplenia syndrome: A review of the relationship with viscero-atrial situs and the spectrum of extra-cardiac anomalies. Surg. Radiol. Anat. 2013, 35, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Rudat, C.; Kispert, A. Wt1 and Epicardial Fate Mapping. Circ. Res. 2012, 111, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-P.; Dong, X.-R.; Regan, J.N.; Su, C.; Majesky, M.W. Tbx18 regulates development of the epicardium and coronary vessels. Dev. Biol. 2013, 383, 307–320. [Google Scholar] [CrossRef] [Green Version]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Sandberg, M.B.; Eskildsen, T.; Nielsen, S.B.; Kalluri, R.; Sheikh, S.P. miR-21 Promotes Fibrogenic Epithelial-to-Mesenchymal Transition of Epicardial Mesothelial Cells Involving Programmed Cell Death 4 and Sprouty-1. PLoS ONE 2013, 8, e56280. [Google Scholar] [CrossRef] [Green Version]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Nossent, A.Y.; Nielsen, S.B.; Sheikh, S.P. Islet-1 is a dual regulator of fibrogenic epithelial-to-mesenchymal transition in epicardial mesothelial cells. Exp. Cell Res. 2013, 319, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Romano, N.; Ceci, M. Heart regeneration is regulates by key micro RNAs from fish to mammals: What it can learned about the epicardial cells activation during the regeneration in zebrafish. Cell Death Dis. 2018, 9, 650. [Google Scholar] [CrossRef] [PubMed]
- Jankowska-Steifer, E.; Niderla-Bielińska, J.; Ciszek, B.; Kujawa, M.; Bartkowiak, M.; Flaht-Zabost, A.; Klosinska, D.; Ratajska, A. Cells with hematopoietic potential reside within mouse proepicardium. Histochem. Cell Biol. 2018, 149, 577–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cossette, S.; Misra, R.P.; Cossett, S. The identification of different endothelial cell populations within the mouse proepicardium. Dev. Dyn. 2011, 240, 2344–2353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilting, J.; Buttler, K.; Schulte, I.; Papoutsi, M.; Schweigerer, L.; Männer, J. The proepicardium delivers hemangioblasts but not lymphangioblasts to the developing heart. Dev. Biol. 2007, 305, 451–459. [Google Scholar] [CrossRef]
- Niderla-Bielińska, J.; Gula, G.; Flaht-Zabost, A.; Jankowska-Steifer, E.; Czarnowska, E.; Radomska-Leśniewska, D.M.; Ciszek, B.; Ratajska, A. 3-D reconstruction and multiple marker analysis of mouse proepicardial endothelial cell population. Microvasc. Res. 2015, 102, 54–69. [Google Scholar] [CrossRef]
- Singh, M.K.; Lu, M.M.; Massera, D.; Epstein, J.A. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J. Biol. Chem. 2011, 286, 41036–41045. [Google Scholar] [CrossRef] [Green Version]
- Dueñas, A.; Expósito, A.; Muñoz, M.D.M.; de Manuel, M.J.; Cámara-Morales, A.; Serrano-Osorio, F.; García-Padilla, C.; Hernández-Torres, F.; Domínguez, J.N.; Aránega, A.; et al. MiR-195 enhances cardiomyogenic differentiation of the proepicardium/septum transversum by Smurf1 and Foxp1 modulation. Sci. Rep. 2020, 10, 9334. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yamagishi, T.; Narematsu, M.; Kamimura, T.; Kai, M.; Nakajima, Y. Epicardium is required for sarcomeric maturation and cardiomyocyte growth in the ventricular compact layer mediated by transforming growth factor β and fibroblast growth factor before the onset of coronary circulation. Congenit Anom. (Kyoto) 2014, 54, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Gittenberger-de Groot, A.C.; Winter, E.M.; Poelmann, R.E. Epicardium-derived cells (EPDCs) in development, cardiac disease and repair of ischemia. J. Cell Mol. Med. 2018, 14, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Tevosian, S.G.; E Deconinck, A.; Tanaka, M.; Schinke, M.; Litovsky, S.H.; Izumo, S.; Fujiwara, Y.; Orkin, S.H. FOG-2, a Cofactor for GATA Transcription Factors, Is Essential for Heart Morphogenesis and Development of Coronary Vessels from Epicardium. Cell 2000, 101, 729–739. [Google Scholar] [CrossRef] [Green Version]
- Towbin, J.A.; Lorts, A.; Jefferies, J.L. Left ventricular non-compaction cardiomyopathy. Lancet 2015, 386, 813–825. [Google Scholar] [CrossRef]
- Weisz, S.H.; Limongelli, G.; Pacileo, G.; Calabrò, P.; Russo, M.G.; Calabrò, R.; Vatta, M. Left Ventricular Non Compaction in Children. Congenit. Heart Dis. 2010, 5, 384–397. [Google Scholar] [CrossRef]
- Ikeda, U.; Minamisawa, M.; Koyama, J. Isolated left ventricular non-compaction cardiomyopathy in adults. J. Cardiol. 2015, 65, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pomares, J.M.; De La Pompa, J.L.; Franco, D.; Henderson, D.; Ho, S.Y.; Houyel, L.; Kelly, R.G.; Sedmera, D.; Sheppard, M.; Sperling, S.; et al. Congenital coronary artery anomalies: A bridge from embryology to anatomy and pathophysiology—A position statement of the development, anatomy, and pathology ESC Working Group. Cardiovasc. Res. 2016, 109, 204–216. [Google Scholar] [CrossRef] [Green Version]
- Moorman, A.F.M.; Christoffels, V.M. Cardiac Chamber Formation: Development, Genes, and Evolution. Physiol. Rev. 2003, 83, 1223–1267. [Google Scholar] [CrossRef]
- Van den Berg, G.; Abu-Issa, R.; de Boer, B.A.; Hutson, M.R.; de Boer, P.A.; Soufan, A.T.; Ruijter, J.M.; Kirby, M.L.; van den Hoff, M.J.; Moorman, A.F. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ. Res. 2009, 104, 179–188. [Google Scholar] [CrossRef]
- Sedmera, D.; Reckova, M.; DeAlmeida, A.; Coppen, S.R.; Kubalak, S.W.; Gourdie, R.G.; Thompson, R.P. Spatiotemporal pattern of commitment to slowed proliferation in the embryonic mouse heart indicates progressive differentiation of the cardiac conduction system. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2003, 274, 773–777. [Google Scholar] [CrossRef]
- Jensen, B.; Wang, T.; Christoffels, V.M.; Moorman, A.F.M. Evolution and development of the building plan of the vertebrate heart. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1833, 783–794. [Google Scholar] [CrossRef] [Green Version]
- Moorman, A.F.M.; Soufan, A.T.; Hagoort, J.; Boer, P.A.J.; Christoffels, V.M. Development of the Building Plan of the Heart. Ann. N. Y. Acad. Sci. 2004, 1015, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, A.; Lozano, E.; Daimi, H.; Esteban, F.J.; Crist, C.; Aranega, A.E.; Franco, D. MicroRNA profiling during mouse ventricular maturation: A role for miR-27 modulating Mef2c expression. Cardiovasc. Res. 2010, 89, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Bonet, F.; Hernández-Torres, F.; Esteban, F.J.; Aranega, A.; Franco, D. Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives. Microarrays 2013, 2, 81–96. [Google Scholar] [CrossRef]
- Qin, X.; Gao, S.; Yang, Y.; Wu, L.; Wang, L. microRNA-25 promotes cardiomyocytes proliferation and migration via targeting Bim. J. Cell. Physiol. 2019, 234, 22103–22115. [Google Scholar] [CrossRef]
- García-Padilla, C.; Domínguez, J.N.; Aránega, A.E.; Franco, D. Differential chamber-specific expression and regulation of long non-coding RNAs during cardiac development. Biochim. Biophys. Acta (BBA)–Bioenerg. 2019, 1862, 194435. [Google Scholar] [CrossRef]
- Pozzo, E.; Chai, Y.C.; Sampaolesi, M. Comprehensive Overview of Non-coding RNAs in Cardiac Development. Adv. Exp. Med. Biol. 2020, 1229, 197–211. [Google Scholar] [CrossRef]
- Deacon, D.C.; Nevis, K.R.; Cashman, T.J.; Zhou, Y.; Zhao, L.; Washko, D.; Guner-Ataman, B.; Burns, C.G.; Burns, C.G.; Burns, C.E. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development 2010, 137, 1887–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morton, S.U.; Scherz, P.J.; Cordes, K.R.; Ivey, K.N.; Stainier, D.Y.R.; Srivastava, D. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. USA 2008, 105, 17830–17835. [Google Scholar] [CrossRef] [Green Version]
- Mosimann, C.; Panáková, D.; Werdich, A.A.; Musso, G.; Burger, A.; Lawson, K.L.; Carr, L.A.; Nevis, K.R.; Sabeh, M.K.; Zhou, Y.; et al. Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 2015, 6, 8146. [Google Scholar] [CrossRef] [Green Version]
- Niessen, K.; Karsan, A. Notch Signaling in Cardiac Development. Circ. Res. 2008, 102, 1169–1181. [Google Scholar] [CrossRef] [Green Version]
- Grego-Bessa, J.; Luna-Zurita, L.; Del Monte, G.; Bolós, V.; Melgar, P.; Arandilla, A.; Garratt, A.N.; Zang, H.; Mukouyama, Y.-S.; Chen, H.; et al. Notch Signaling Is Essential for Ventricular Chamber Development. Dev. Cell 2007, 12, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Samsa, L.A.; Givens, C.; Tzima, E.; Stainier, D.Y.R.; Qian, L.; Liu, J. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development 2015, 142, 4080–4091. [Google Scholar] [CrossRef] [Green Version]
- Del Monte-Nieto, G.; Ramialison, M.; Adam, A.A.S.; Wu, B.; Aharonov, A.; D’Uva, G.; Bourke, L.M.; Pitulescu, M.E.; Chen, H.; de la Pompa, J.L.; et al. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature 2018, 557, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Peshkovsky, C.; Totong, R.; Yelon, D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev. Dyn. 2011, 240, 446–456. [Google Scholar] [CrossRef]
- Lai, D.; Liu, X.; Forrai, A.; Wolstein, O.; Michalicek, J.; Ahmed, I.; Garratt, A.N.; Birchmeier, C.; Zhou, M.; Hartley, L.; et al. Neuregulin 1 Sustains the Gene Regulatory Network in Both Trabecular and Nontrabecular Myocardium. Circ. Res. 2010, 107, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Zhang, X.; Zhao, Q.; Lou, X. The zinc finger protein Zfpm1 modulates ventricular trabeculation through Neuregulin-ErbB signalling. Dev. Biol. 2019, 446, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, S.J.; Stainier, D.Y. Regulation of cardiomyocyte behavior in zebrafish trabeculation by Neuregulin 2a signaling. Nat. Commun. 2017, 8, 15281. [Google Scholar] [CrossRef] [Green Version]
- Negro, A.; Brar, B.K.; Lee, K.-F. Essential roles of Her2/erbB2 in cardiac development and function. Recent Prog. Horm. Res. 2004, 59, 1–12. [Google Scholar] [CrossRef] [Green Version]
- van Weerd, J.H.; Christoffels, V.M. The formation and function of the cardiac conduction system. Development 2016, 143, 197–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, R.A.; Boukens, B.J.; Christoffels, V.M. Developmental Origin of the Cardiac Conduction System: Insight from Lineage Tracing. Pediatr. Cardiol. 2018, 39, 1107–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Eif, V.W.W.; Devalla, H.D.; Boink, G.J.J.; Christoffels, V.M. Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol. 2018, 15, 617–630. [Google Scholar] [CrossRef]
- van Eif, V.W.W.; Stefanovic, S.; Mohan, R.A.; Christoffels, V.M. Gradual differentiation and confinement of the cardiac conduction system as indicated by marker gene expression. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118509. [Google Scholar] [CrossRef]
- Jensen, B.; Boukens, B.J.; Crossley, D.A.; Conner, J.; Mohan, R.A.; van Duijvenboden, K.; Postma, A.V.; Gloschat, C.R.; Elsey, R.M.; Sedmera, D.; et al. Specialized impulse conduction pathway in the alligator heart. eLife 2018, 7, e32120. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B.; Boukens, B.J.D.; Postma, A.V.; Gunst, Q.D.; van den Hoff, M.J.; Moorman, A.F.M.; Wang, T.; Christoffels, V.M. Identifying the Evolutionary Building Blocks of the Cardiac Conduction System. PLoS ONE 2012, 7, e44231. [Google Scholar] [CrossRef] [Green Version]
- Christoffels, V.M.; Smits, G.J.; Kispert, A.; Moorman, A.F.M. Development of the Pacemaker Tissues of the Heart. Circ. Res. 2010, 106, 240–254. [Google Scholar] [CrossRef]
- Bakker, M.L.; Moorman, A.F.M.; Christoffels, V.M. The Atrioventricular Node: Origin, Development, and Genetic Program. Trends Cardiovasc. Med. 2010, 20, 164–171. [Google Scholar] [CrossRef]
- Bakker, M.L.; Christoffels, V.M.; Moorman, A.F.M. The Cardiac Pacemaker and Conduction System Develops From Embryonic Myocardium that Retains Its Primitive Phenotype. J. Cardiovasc. Pharmacol. 2010, 56, 6–15. [Google Scholar] [CrossRef]
- Tessadori, F.; Van Weerd, J.H.; Burkhard, S.B.; Verkerk, A.O.; De Pater, E.; Boukens, B.J.; Vink, A.; Christoffels, V.M.; Bakkers, J. Identification and Functional Characterization of Cardiac Pacemaker Cells in Zebrafish. PLoS ONE 2012, 7, e47644. [Google Scholar] [CrossRef] [Green Version]
- Kvasilova, A.; Olejnickova, V.; Jensen, B.; Christoffels, V.M.; Kolesova, H.; Sedmera, D.; Gregorovicova, M. The formation of the atrioventricular conduction axis is linked in development to ventricular septation. J. Exp. Biol. 2020, 223, jeb229278. [Google Scholar] [CrossRef]
- Aanhaanen, W.T.J.; Mommersteeg, M.T.M.; Norden, J.; Wakker, V.; Vries, C.D.G.-D.; Anderson, R.H.; Kispert, A.; Moorman, A.F.M.; Christoffels, V.M. Developmental Origin, Growth, and Three-Dimensional Architecture of the Atrioventricular Conduction Axis of the Mouse Heart. Circ. Res. 2010, 107, 728–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicente-Steijn, R.; Kelder, T.P.; Tertoolen, L.G.; Wisse, L.J.; Pijnappels, D.A.; Poelmann, R.; Schalij, M.J.; DeRuiter, M.C.; Groot, A.C.G.-D.; Jongbloed, M. RHOA-ROCK signalling is necessary for lateralization and differentiation of the developing sinoatrial node. Cardiovasc. Res. 2017, 113, 1186–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourdie, R.G.; Mima, T.; Thompson, R.P.; Mikawa, T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development 1995, 121, 1423–1431. [Google Scholar]
- Gourdie, R.G.; Wei, Y.; Kim, D.; Klatt, S.C.; Mikawa, T. Endothelin-induced conversion of embryonic heart muscle cells into impulse-conducting Purkinje fibers. Proc. Natl. Acad. Sci. USA 1998, 95, 6815–6818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourdie, R.G.; Harris, B.S.; Bond, J.; Edmondson, A.M.; Cheng, G.; Sedmera, D.; O’Brien, T.X.; Mikawa, T.; Thompson, R.P. His-Purkinje lineages and development. Novartis Found. Symp. 2003, 250, 110–124. [Google Scholar] [CrossRef]
- Gourdie, R.G.; Harris, B.S.; Bond, J.; Justus, C.; Hewett, K.W.; O’Brien, T.X.; Thompson, R.P.; Sedmera, D. Development of the cardiac pacemaking and conduction system. Birth Defects Res. C Embryo Today 2003, 69, 46–57. [Google Scholar] [CrossRef]
- Cheng, G.; Litchenberg, W.H.; Cole, G.J.; Mikawa, T.; Thompson, R.P.; Gourdie, R.G. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development 1999, 126, 5041–5049. [Google Scholar]
- Sedmera, D.; Harris, B.S.; Grant, E.; Zhang, N.; Jourdan, J.; Kurková, D.; Gourdie, R.G. Cardiac expression patterns of endothelin-converting enzyme (ECE): Implications for conduction system development. Dev. Dyn. 2008, 237, 1746–1753. [Google Scholar] [CrossRef] [Green Version]
- Takebayashi-Suzuki, K.; Yanagisawa, M.; Gourdie, R.G.; Kanzawa, N.; Mikawa, T. In vivo induction of cardiac Purkinje fiber differentiation by coexpression of preproendothelin-1 and endothelin converting enzyme-1. Development 2000, 127, 3523–3532. [Google Scholar]
- Hyer, J.; Johansen, M.; Prasad, A.; Wessels, A.; Kirby, M.L.; Gourdie, R.G.; Mikawa, T. Induction of Purkinje fiber differentiation by coronary arterialization. Proc. Natl. Acad. Sci. USA 1999, 96, 13214–13218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miquerol, L.; Moreno-Rascon, N.; Beyer, S.; Dupays, L.; Meilhac, S.M.; Buckingham, M.E.; Franco, D.; Kelly, R.G. Biphasic Development of the Mammalian Ventricular Conduction System. Circ. Res. 2010, 107, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Miquerol, L.; Beyer, S.; Kelly, R.G. Establishment of the mouse ventricular conduction system. Cardiovasc. Res. 2011, 91, 232–242. [Google Scholar] [CrossRef] [Green Version]
- Miquerol, L.; Bellon, A.; Moreno, N.; Beyer, S.; Meilhac, S.M.; Buckingham, M.; Franco, D.; Kelly, R.G. Resolving cell lineage contributions to the ventricular conduction system with a Cx40-GFP allele: A dual contribution of the first and second heart fields. Dev. Dyn. 2013, 242, 665–677. [Google Scholar] [CrossRef]
- Choquet, C.; Kelly, R.G.; Miquerol, L. Defects in Trabecular Development Contribute to Left Ventricular Noncompaction. Pediatr. Cardiol. 2019, 40, 1331–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choquet, C.; Marcadet, L.; Beyer, S.; Kelly, R.G.; Miquerol, L. Segregation of Central Ventricular Conduction System Lineages in Early SMA+ Cardiomyocytes Occurs Prior to Heart Tube Formation. J. Cardiovasc. Dev. Dis. 2016, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Choquet, C.; Nguyen, T.H.M.; Sicard, P.; Buttigieg, E.; Tran, T.T.; Kober, F.; Varlet, I.; Sturny, R.; Costa, M.W.; Harvey, R.P.; et al. Deletion of Nkx2-5 in trabecular myocardium reveals the developmental origins of pathological heterogeneity associated with ventricular non-compaction cardiomyopathy. PLoS Genet. 2018, 14, e1007502. [Google Scholar]
- Bakker, M.L.; Boukens, B.J.; Mommersteeg, M.T.M.; Brons, J.F.; Wakker, V.; Moorman, A.F.M.; Christoffels, V.M. Transcription Factor Tbx3 Is Required for the Specification of the Atrioventricular Conduction System. Circ. Res. 2008, 102, 1340–1349. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Bai, Y.; Li, N.; Ye, W.; Zhang, M.; Greene, S.B.; Tao, Y.; Chen, Y.; Wehrens, X.H.T.; Martin, J.F. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA 2014, 111, 9181–9186. [Google Scholar] [CrossRef] [Green Version]
- Bond, J.; Sedmera, D.; Jourdan, J.; Zhang, Y.; Eisenberg, C.A.; Eisenberg, L.M.; Gourdie, R.G. Wnt11 and Wnt7a are up-regulated in association with differentiation of cardiac conduction cells in vitro and in vivo. Dev. Dyn. 2003, 227, 536–543. [Google Scholar] [CrossRef]
- Samal, E.; Evangelista, M.; Galang, G.; Srivastava, D.; Zhao, Y.; Vedantham, V. Premature MicroRNA-1 Expression Causes Hypoplasia of the Cardiac Ventricular Conduction System. Front. Physiol. 2019, 10, 235. [Google Scholar] [CrossRef]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; Von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef] [Green Version]
- Garnatz, A.S.; Gao, Z.; Broman, M.; Martens, S.; Earley, J.U.; Svensson, E.C. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev. Biol. 2014, 395, 50–61. [Google Scholar] [CrossRef] [Green Version]
- Shamis, Y.; Cullen, D.E.; Liu, L.; Yang, G.; Ng, S.F.; Xiao, L.; Bell, F.T.; Ray, C.; Takikawa, S.; Moskowitz, I.P.; et al. Maternal and zygotic Zfp57 modulate NOTCH signaling in cardiac development. Proc. Natl. Acad. Sci. USA 2015, 112, E2020–E2029. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Qin, L.; Liu, Z.; Liao, L.; Martin, J.F.; Xu, J. Knockout of SRC-1 and SRC-3 in Mice Decreases Cardiomyocyte Proliferation and Causes a Noncompaction Cardiomyopathy Phenotype. Int. J. Biol. Sci. 2015, 11, 1056–1072. [Google Scholar] [CrossRef] [Green Version]
- Harmelink, C.; Peng, Y.; DeBenedittis, P.; Chen, H.; Shou, W.; Jiao, K. Myocardial Mycn is essential for mouse ventricular wall morphogenesis. Dev. Biol. 2013, 373, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Xiuzhen, H.; Tian, X.; Zhang, H.; Hu, T.; Huang, X.; Zhang, L.; Wang, Z.; Zhou, B. BAF200 Is Required for Heart Morphogenesis and Coronary Artery Development. PLoS ONE 2014, 9, e109493. [Google Scholar] [CrossRef] [Green Version]
- Yagi, H.; Liu, X.; Gabriel, G.C.; Wu, Y.; Peterson, K.; Murray, S.A.; Aronow, B.J.; Martin, L.J.; Benson, D.W.; Lo, C.W. The Genetic Landscape of Hypoplastic Left Heart Syndrome. Pediatr. Cardiol. 2018, 39, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yagi, H.; Saeed, S.; Bais, A.S.; Gabriel, G.C.; Chen, Z.; A Peterson, K.; Li, Y.; Schwartz, M.C.; Reynolds, W.T.; et al. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017, 49, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, B.G.; Bao, Z.-Z.; Fatkin, D.; Xavier-Neto, J.; Georgakopoulos, D.; Maguire, C.T.; Berul, C.I.; Kass, D.A.; Bold, M.L.K.-D.; De Bold, A.J.; et al. Cardiomyopathy in Irx4-Deficient Mice Is Preceded by Abnormal Ventricular Gene Expression. Mol. Cell. Biol. 2001, 21, 1730–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomita-Mitchell, A.; Stamm, K.D.; Mahnke, D.K.; Kim, M.-S.; Hidestrand, P.M.; Liang, H.L.; Goetsch, M.A.; Hidestrand, M.; Simpson, P.; Pelech, A.N.; et al. Impact of MYH6 variants in hypoplastic left heart syndrome. Physiol. Genom. 2016, 48, 912–921. [Google Scholar] [CrossRef] [Green Version]
- Durbin, M.D.; Cadar, A.G.; Williams, C.H.; Guo, Y.; Bichell, D.P.; Su, Y.R.; Hong, C.C. Hypoplastic Left Heart Syndrome Sequencing Reveals a Novel NOTCH1 Mutation in a Family with Single Ventricle Defects. Pediatr. Cardiol. 2017, 38, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.K.; Bhat, A.M.; Conard, K.; Hyland, J.; Pizarro, C. Novel SMAD3 Mutation in a Patient with Hypoplastic Left Heart Syndrome with Significant Aortic Aneurysm. Case Rep. Genet. 2014, 2014, 591516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhagen, J.M.; Born, M.V.D.; Kurul, S.; Asimaki, A.; Van De Laar, I.M.B.H.; Frohn-Mulder, I.M.; Kammeraad, J.A.; Yap, S.C.; Bartelings, M.M.; Van Slegtenhorst, M.A.; et al. Homozygous Truncating Variant in PKP2 Causes Hypoplastic Left Heart Syndrome. Circ. Genom. Precis. Med. 2018, 11, e002397. [Google Scholar] [CrossRef] [Green Version]
- Dueñas, A.; Expósito-Villén, A.; Aranega, A.; Franco, D. The Role of Non-Coding RNA in Congenital Heart Diseases. J. Cardiovasc. Dev. Dis. 2019, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Liang, D.; Xu, X.; Deng, F.; Feng, J.; Zhang, H.; Liu, Y.; Zhang, Y.; Pan, L.; Liu, Y.; Zhang, D.; et al. miRNA-940 reduction contributes to human Tetralogy of Fallot development. J. Cell Mol. Med. 2014, 18, 1830–1839. [Google Scholar] [CrossRef]
- Saxena, A.; Tabin, C.J. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc. Natl. Acad. Sci. USA. 2010, 107, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Hu, Z.; Xu, Z.; Gu, H.; Yi, L.; Cao, H.; Chen, J.; Tian, T.; Liang, J.; Lin, Y.; et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum. Mutat. 2009, 30, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Yoshida, M.; Tarui, S.; Hirata, M.; Nagai, Y.; Kasahara, S.; Naruse, K.; Ito, H.; Sano, S.; Oh, H. Directed Differentiation of Patient-Specific Induced Pluripotent Stem Cells Identifies the Transcriptional Repression and Epigenetic Modification of NKX2-5, HAND1, and NOTCH1 in Hypoplastic Left Heart Syndrome. PLoS ONE 2014, 9, e102796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aanhaanen, W.T.; Boukens, B.J.; Sizarov, A.; Wakker, V.; Vries, C.D.G.-D.; Van Ginneken, A.C.; Moorman, A.F.; Coronel, R.; Christoffels, V.M. Defective Tbx2-dependent patterning of the atrioventricular canal myocardium causes accessory pathway formation in mice. J. Clin. Investig. 2011, 121, 534–544. [Google Scholar] [CrossRef] [Green Version]
- Frank, D.U.; Carter, K.L.; Thomas, K.R.; Burr, R.M.; Bakker, M.L.; Coetzee, W.A.; Tristani-Firouzi, M.; Bamshad, M.J.; Christoffels, V.M.; Moon, A.M. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc. Natl. Acad. Sci. USA 2011, 109, E154–E163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theis, J.L.; Zimmermann, M.T.; Evans, J.M.; Eckloff, B.W.; Wieben, E.D.; Qureshi, M.Y.; O’Leary, P.W.; Olson, T.M. Recessive MYH6 Mutations in Hypoplastic Left Heart With Reduced Ejection Fraction. Circ. Cardiovasc. Genet. 2015, 8, 564–571. [Google Scholar] [CrossRef] [Green Version]
- Theis, J.L.; Hrstka, S.C.L.; Evans, J.M.; O’Byrne, M.M.; De Andrade, M.; O’Leary, P.W.; Nelson, T.J.; Olson, T.M. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Qual. Life Res. 2015, 134, 1003–1011. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Wei, C.; Hou, Z.; Li, Y.; Zou, H.; Meng, M.; Wang, W.; Jiang, L. Genetic variations of NKX2-5 in sporadic atrial septal defect and ventricular septal defect in Chinese Yunnan population. Gene 2016, 575, 29–33. [Google Scholar] [CrossRef]
- Hirayama-Yamada, K.; Kamisago, M.; Akimoto, K.; Aotsuka, H.; Nakamura, Y.; Tomita, H.; Furutani, M.; Imamura, S.-I.; Takao, A.; Nakazawa, M.; et al. Phenotypes withGATA4 orNKX2.5 mutations in familial atrial septal defect. Am. J. Med. Genet. Part A 2005, 135, 47–52. [Google Scholar] [CrossRef]
- Mohan, R.A.; Van Engelen, K.; Stefanovic, S.; Barnett, P.; Ilgun, A.; Baars, M.J.H.; Bouma, B.J.; Mulder, B.J.M.; Christoffels, V.M.; Postma, A.V. A mutation in the Kozak sequence ofGATA4hampers translation in a family with atrial septal defects. Am. J. Med. Genet. Part A 2014, 164, 2732–2738. [Google Scholar] [CrossRef]
- Xu, Y.-J.; Qiu, X.-B.; Yuan, F.; Shi, H.-Y.; Xu, L.; Hou, X.-M.; Qu, X.-K.; Liu, X.; Huang, R.-T.; Xue, S.; et al. Prevalence and spectrum of NKX2.5 mutations in patients with congenital atrial septal defect and atrioventricular block. Mol. Med. Rep. 2017, 15, 2247–2254. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Qi, B.; Zhao, J.; Liu, W.; Duan, R.; Zhang, M. A novel mutation of GATA4 (K300T) associated with familial atrial septal defect. Gene 2016, 575, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Fanm, L.L.; Huang, H.; Cao, B.B.; Li, X.P.; Peng, D.Q.; Xia, K. A novel mutation of GATA4 (K319E) is responsible for familial atrial septal defect and pulmonary valve stenosis. Gene 2014, 534, 320–323. [Google Scholar] [CrossRef]
- Zhou, Y.; Dai, X.; Huang, R.-T.; Xue, S.; Xu, Y.-J.; Qiu, X.-B.; Yang, Y.-Q. A novel TBX20 loss-of-function mutation contributes to adult-onset dilated cardiomyopathy or congenital atrial septal defect. Mol. Med. Rep. 2016, 14, 3307–3314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.-M.; Wang, J.; Qiu, X.-B.; Yuan, F.; Li, R.-G.; Xu, Y.-J.; Qu, X.-K.; Shi, H.-Y.; Hou, X.-M.; Huang, R.-T.; et al. A HAND2 Loss-of-Function Mutation Causes Familial Ventricular Septal Defect and Pulmonary Stenosis. G3 (Bethesda) 2016, 6, 987–992. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Mao, J.-H.; Ding, K.-K.; Xu, W.-J.; Liu, X.-Y.; Qiu, X.-B.; Li, R.-G.; Qu, X.-K.; Xu, Y.-J.; Huang, R.-T.; et al. A Novel NKX2.6 Mutation Associated with Congenital Ventricular Septal Defect. Pediatr. Cardiol. 2014, 36, 646–656. [Google Scholar] [CrossRef]
- Winston, J.B.; Schulkey, C.E.; Chen, I.-B.D.; Regmi, S.D.; Efimova, M.; Erlich, J.M.; Green, C.A.; Aluko, A.; Jay, P.Y. Complex Trait Analysis of Ventricular Septal Defects Caused by Nkx2-5 Mutation. Circ. Cardiovasc. Genet. 2012, 5, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Li, B.; Li, F.; Sun, K.; Jiang, X.; Xu, R. Identification of FOXH1 mutations in patients with sporadic conotruncal heart defect. Clin. Genet. 2020, 97, 576–585. [Google Scholar] [CrossRef]
- Zhao, C.-M.; Peng, L.-Y.; Li, L.; Liu, X.-Y.; Wang, J.; Zhang, X.-L.; Yuan, F.; Li, R.-G.; Qiu, X.-B.; Yang, Y.-Q. PITX2 Loss-of-Function Mutation Contributes to Congenital Endocardial Cushion Defect and Axenfeld-Rieger Syndrome. PLoS ONE 2015, 10, e0124409. [Google Scholar] [CrossRef] [Green Version]
- Zhang, E.; Yang, J.; Liu, Y.; Hong, N.; Xie, H.; Fu, Q.; Li, F.; Chen, S.; Yu, Y.; Sun, K. MESP2 variants contribute to conotruncal heart defects by inhibiting cardiac neural crest cell proliferation. J. Mol. Med. 2020, 98, 1035–1048. [Google Scholar] [CrossRef]
- Zhang, M.; Li, F.X.; Liu, X.Y.; Huang, R.T.; Xue, S.; Yang, X.X.; Li, Y.J.; Liu, H.; Shi, H.Y.; Pan, X.; et al. MESP1 loss‑of‑function mutation contributes to double outlet right ventricle. Mol. Med. Rep. 2017, 16, 2747–2754. [Google Scholar] [CrossRef]
- Lin, X.; Huo, Z.; Liu, X.; Zhang, Y.; Li, L.; Zhao, H.; Yan, B.; Liu, Y.; Yang, Y.; Chen, Y.-H. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J. Hum. Genet. 2010, 55, 662–667. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.-X.; Gong, H.-R.; Liu, X.; Wang, J.; Zhao, C.-M.; Huang, R.-T.; Xue, S.; Yang, Y.-Q. A novel HAND2 loss-of-function mutation responsible for tetralogy of Fallot. Int. J. Mol. Med. 2016, 37, 445–451. [Google Scholar] [CrossRef] [Green Version]
- Li, R.-G.; Li, L.; Qiu, X.; Yuan, F.; Xu, L.; Li, X.; Xu, Y.; Jiang, W.-F.; Jiang, J.-Q.; Liu, X.; et al. GATA4 loss-of-function mutation underlies familial dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2013, 439, 591–596. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, W.-D.; Yang, Z.-L.; Yuan, F.; Xu, L.; Li, R.-G.; Yang, Y.-Q. Prevalence and spectrum of GATA4 mutations associated with sporadic dilated cardiomyopathy. Gene 2014, 548, 174–181. [Google Scholar] [CrossRef]
- Zhao, L.; Xu, J.-H.; Xu, W.-J.; Yu, H.; Wang, Q.; Zheng, H.-Z.; Jiang, W.-F.; Jiang, J.-F.; Yang, Y.-Q. A novel GATA4 loss-of-function mutation responsible for familial dilated cardiomyopathy. Int. J. Mol. Med. 2013, 33, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Qiu, X.-B.; Yuan, F.; Wang, J.; Zhao, C.-M.; Li, R.-G.; Xu, L.; Xu, Y.-J.; Shi, H.; Hou, X.-M.; et al. TBX5 loss-of-function mutation contributes to familial dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2015, 459, 166–171. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Zhao, L.; Jiang, J.-Q.; Jiang, W.-F.; Yang, Y.-Q.; Qiu, X.-B. A novel TBX5 loss-of-function mutation associated with sporadic dilated cardiomyopathy. Int. J. Mol. Med. 2015, 36, 282–288. [Google Scholar] [CrossRef]
- Xu, J.-H.; Gu, J.-Y.; Guo, Y.-H.; Zhang, H.; Qiu, X.-B.; Li, R.-G.; Shi, H.-Y.; Liu, H.; Yang, X.-X.; Xu, Y.-J.; et al. Prevalence and Spectrum of NKX2-5 Mutations Associated With Sporadic Adult-Onset Dilated Cardiomyopathy. Int. Heart J. 2017, 58, 521–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, F.; Qiu, Z.-H.; Wang, X.-H.; Sun, Y.-M.; Wang, J.; Li, R.-G.; Liu, H.; Zhang, M.; Shi, H.-Y.; Zhao, L.; et al. MEF2C loss-of-function mutation associated with familial dilated cardiomyopathy. Clin. Chem. Lab. Med. 2017, 56, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Li, R.-G.; Xu, Y.; Wang, J.; Liu, X.; Yuan, F.; Huang, R.-T.; Xue, S.; Li, L.; Liu, H.; Li, Y.-J.; et al. GATA4 Loss-of-Function Mutation and the Congenitally Bicuspid Aortic Valve. Am. J. Cardiol. 2018, 121, 469–474. [Google Scholar] [CrossRef]
- Qu, X.-K.; Qiu, X.-B.; Yuan, F.; Wang, J.; Zhao, C.-M.; Liu, X.-Y.; Zhang, X.-L.; Li, R.-G.; Xu, Y.-J.; Hou, X.-M.; et al. A Novel NKX2.5 Loss-of-Function Mutation Associated With Congenital Bicuspid Aortic Valve. Am. J. Cardiol. 2014, 114, 1891–1895. [Google Scholar] [CrossRef]
- Posch, M.G.; Boldt, L.-H.; Polotzki, M.; Richter, S.; Rolf, S.; Perrot, A.; Dietz, R.; Özcelik, C.; Haverkamp, W. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur. J. Med. Genet. 2010, 53, 201–203. [Google Scholar] [CrossRef]
- Yuan, F.; Qiu, X.-B.; Li, R.-G.; Qu, X.-K.; Wang, J.; Xu, Y.-J.; Liu, X.; Fang, W.-Y.; Yang, Y.-Q.; Liao, D.-N. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int. J. Mol. Med. 2014, 35, 478–486. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.-F.; Yang, F.; Mahida, S.N.; Zhao, L.; Chen, X.; Zhang, M.L.; Sun, Z.; Yao, Y.; Zhang, Y.-X.; Zheng, G.-Y.; et al. TBX5 mutations contribute to early-onset atrial fibrillation in Chinese and Caucasians. Cardiovasc. Res. 2016, 109, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zhou, L.; Wang, Q.; You, X.; Li, Y.; Zhao, Y.; Han, X.; Chang, Z.; He, X.; Cheng, C.; et al. NEXN inhibits GATA4 and leads to atrial septal defects in mice and humans. Cardiovasc. Res. 2014, 103, 228–237. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Feliciano, J.; Lee, K.-H.; Kong, S.W.; Rajagopal, S.; Ma, Q.; Springer, Z.; Izumo, S.; Tabin, C.J.; Pu, W.T. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 2006, 133, 3607–3618. [Google Scholar] [CrossRef] [Green Version]
- de Vlaming, A.; Sauls, K.; Hajdu, Z.; Visconti, R.P.; Mehesz, A.N.; Levine, R.A.; Slaugenhaupt, S.A.; Hagège, A.; Chester, A.H.; Markwald, R.R.; et al. Atrioventricular valve development: New perspectives on an old theme. Differentiation 2012, 84, 103–116. [Google Scholar] [CrossRef] [Green Version]
- Stefanovic, S.; Barnett, P.; Van Duijvenboden, K.; Weber, D.; Gessler, M.; Christoffels, V.M. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat. Commun. 2014, 5, 3680. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, M.M.; Wirrig, E.E.; Phelps, A.L.; Ghatnekar, A.V.; Barth, J.L.; Norris, R.A.; Wessels, A. Mef2c Regulates Transcription of the Extracellular Matrix Protein Cartilage Link Protein 1 in the Developing Murine Heart. PLoS ONE 2013, 8, e57073. [Google Scholar] [CrossRef] [Green Version]
- Franco, D.; Meilhac, S.M.; Christoffels, V.M.; Kispert, A.; Buckingham, M.; Kelly, R.G. Left and right ventricular contributions to the formation of the interventricular septum in the mouse heart. Dev. Biol. 2006, 294, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, J.K.; Ohgi, M.; Koshiba-Takeuchi, K.; Shiratori, H.; Sakaki, I.; Ogura, T.; Saijoh, Y. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development 2003, 130, 5953–5964. [Google Scholar] [CrossRef] [Green Version]
- Vanyai, H.K.; Thomas, T.; Voss, A.K. Mesodermal expression of Moz is necessary for cardiac septum development. Dev. Biol. 2015, 403, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Yuasa, S.; Onizuka, T.; Shimoji, K.; Ohno, Y.; Kageyama, T.; Yoon, S.H.; Egashira, T.; Seki, T.; Hashimoto, H.; Nishiyama, T.; et al. Zac1 Is an Essential Transcription Factor for Cardiac Morphogenesis. Circ. Res. 2010, 106, 1083–1091. [Google Scholar] [CrossRef] [Green Version]
- Panzer, A.A.; Regmi, S.D.; Cormier, D.; Danzo, M.T.; Chen, I.-B.D.; Winston, J.B.; Hutchinson, A.K.; Salm, D.; Schulkey, C.E.; Cochran, R.S.; et al. Nkx2-5 and Sarcospan genetically interact in the development of the muscular ventricular septum of the heart. Sci. Rep. 2017, 7, 46438. [Google Scholar] [CrossRef] [Green Version]
- Ya, J.; van den Hoff, M.J.; de Boer, P.A.; Tesink-Taekema, S.; Franco, D.; Moorman, A.F.; Lamers, W.H. Normal development of the outflow tract in the rat. Circ Res. 1998, 82, 464–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, D.; Markman, M.M.; Wagenaar, G.T.; Ya, J.; Lamers, W.H.; Moorman, A.F. Myosin light chain 2a and 2v identifies the embryonic outflow tract myocardium in the developing rodent heart. Anat. Rec. 1999, 254, 135–146. [Google Scholar] [CrossRef]
- Miyagawa-Tomita, S.; Waldo, K.; Tomita, H.; Kirby, M.L. Temporospatial study of the migration and distribution of cardiac neural crest in quail-chick chimeras. Am. J. Anat. 1991, 192, 79–88. [Google Scholar] [CrossRef]
- Kirby, M.L. Cellular and molecular contributions of the cardiac neural crest to cardiovascular development. Trends Cardiovasc. Med. 1993, 3, 18–23. [Google Scholar] [CrossRef]
- Creazzo, T.L.; Godt, R.E.; Leatherbury, L.; Conway, S.J.; Kirby, M.L. Role of cardiac neural crest cells in cardiovascular development. Annu. Rev. Physiol. 1998, 60, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Kirby, M.L.; Turnage, K.L.; Hays, B.M. Characterization of conotruncal malformations following ablation of “cardiac” neural crest. Anat. Rec. 1985, 213, 87–93. [Google Scholar] [CrossRef]
- Bookman, D.E.; Redmond, M.E.; Waldo, K.; Davis, H.; Kirby, M.L. Effect of neural crest ablation on development of the heart and arch arteries in the chick. Am. J. Anat. 1987, 180, 332–341. [Google Scholar] [CrossRef]
- Yelbuz, T.M.; Waldo, K.L.; Kumiski, D.H.; Stadt, H.A.; Wolfe, R.R.; Leatherbury, L.; Kirby, M.L. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation 2002, 106, 504–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldo, K.; Miyagawa-Tomita, S.; Kumiski, D.; Kirby, M.L. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: Aortic sac to ventricular septal closure. Dev. Biol. 1998, 196, 129–144. [Google Scholar] [CrossRef]
- Kanne, J.P.; Godwin, J.D. Right aortic arch and its variants. J. Cardiovasc. Comput. Tomogr. 2010, 4, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Gobel, J.W.; Pierpont, M.E.M.; Moller, J.H.; Singh, A.; Edwards, J.E. Familial interruption of the aortic arch. Pediatr. Cardiol. 1993, 14, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Bockman, D.E.; Redmond, M.E.; Kirby, M.L. Alteration of early vascular development after ablation of cranial neural crest. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 1989, 225, 209–217. [Google Scholar] [CrossRef]
- Kurihara, Y.; Kurihara, H.; Oda, H.; Maemura, K.; Nagai, R.; Ishikawa, T.; Yazaki, Y. Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J. Clin. Investig. 1995, 96, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, H.; Hammer, R.E.; Richardson, J.A.; Williams, S.C.; Clouthier, D.E.; Yanagisawa, M. Role of Endothelin-1/Endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J. Clin. Investig. 1998, 102, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Thomas, T.; Kurihara, H.; Yamagishi, H.; Kurihara, Y.; Yazaki, Y.; Olson, E.N.; Srivastava, D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development 1998, 125, 3005–3014. [Google Scholar] [PubMed]
- Sugrue, K.F.; Sarkar, A.A.; Leatherbury, L.; Zohn, I.E. The ubiquitin ligase HECTD1 promotes retinoic acid signaling required for development of the aortic arch. Dis. Model. Mech. 2019, 12, dmm036491. [Google Scholar] [CrossRef] [Green Version]
- Niederreither, K.; Vermot, J.; Le Roux, I.; Schuhbaur, B.; Chambon, P.; Dollé, P. The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development 2003, 130, 2525–2534. [Google Scholar] [CrossRef] [Green Version]
- Colbert, M.C.; Kirby, M.L.; Robbins, J. Endogenous retinoic acid signaling colocalizes with advanced expression of the adult smooth muscle myosin heavy chain isoform during development of the ductus arteriosus. Circ. Res. 1996, 78, 790–798. [Google Scholar] [CrossRef]
- Sheehy, N.T.; Cordes, K.R.; White, M.P.; Ivey, K.N.; Srivastava, D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development 2010, 137, 4307–4316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escot, S.; Blavet, C.; Haertle, S.; Duband, J.-L.; Fournier-Thibault, C. Misregulation of SDF1-CXCR4 Signaling Impairs Early Cardiac Neural Crest Cell Migration Leading to Conotruncal Defects. Circ. Res. 2013, 113, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a Long Noncoding RNA Required for Cardiovascular Lineage Commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Müller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657.e8. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1 Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146, dev176198. [Google Scholar] [CrossRef] [Green Version]
- Smart, N.; Bollini, S.; Dubé, K.N.; Vieira, J.M.; Zhou, B.; Davidson, S.; Yellon, D.; Riegler, J.; Price, A.N.; Lythgoe, M.F.; et al. De novo cardiomyocytes from within the activated adult heart after injury. Nat. Cell Biol. 2011, 474, 640–644. [Google Scholar] [CrossRef] [Green Version]



| TF | Function | References |
|---|---|---|
| Brachyury | Mesodermal commitment | [1] |
| Mesp1 | Cardiogenic mesoderm commitment | [2,3,4,5,6,7,8,9] |
| Mesp2 | Cardiogenic mesoderm commitment | [2,3,4,5,6,7,8,9] |
| Gata4 | Early cardiac specification, proepicardium development, chamber formation, atrial and atrioventricular septation, cardiomyocyte proliferation, cardiac hypertrophy | [10,11,12,13,14,27,35,36,37,38,94,95,97,98,99,100,125,126,127] |
| Gata5 | Early cardiac specification, cardiomyocyte proliferation | [10,11,12,13,14,16,26,105] |
| Gata6 | Early cardiac specification, cardiac hypertrophy | [10,11,12,13,14,16,25,41] |
| Nkx2.3 | Early cardiac specification | [15] |
| Nkx2.5 | Early cardiac specification, FHF development, cardiac looping, chamber formation, cardiac conduction system specification, atrial septation | [15,31,32,33,34,68,96,97,98,99,100,128,129,130,131,132,133,134,135] |
| Nkx2.6 | Early cardiac specification | [24,29] |
| Nkx2.7 | Early cardiac specification | [15] |
| Nkx2.8 | Early cardiac specification | [18,19,20] |
| Foxh1 | Anterior heart field development | [28,35] |
| Mef2c | Early cardiac specification, chamber formation | [35,36,37,38,39,40,41,42,43,136] |
| Islet-1 | Second heart field specification | [69] |
| Tbx5 | Early cardiac specification, chamber formation, cardiac conduction system specification, atrial and ventricular septation | [101,102,103,104,128,137,138,139,140,141,142] |
| Pitx2 | left right signalling, heterotaxia, chamber formation, cardiac conduction system development | [143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167] |
| Prrx1 | cardiac looping | [168] |
| Wt1 | proepicardium and epicardial development | [169,170] |
| Tcf21 | proepicardium and epicardial development | [171,172] |
| Tbx18 | proepicardium and epicardial development | [169] |
| Tbx2 | primary myocardium and cardiac conduction system development | [173,174,175,176,177,178] |
| Tbx3 | primary myocardium and cardiac conduction system development | [179,180,181,182] |
| Tbx20 | chamber formation, atrioventricular canal development, cardiomyocyte cell proliferation | [183,184,185,186,187,188,189,190] |
| Irx3 | ventricular trabecular and cardiac conduction system development | [191,192] |
| Irx4 | ventricular chamber development | [191,193] |
| Irx5 | cardiac conduction system development | [191,192] |
| Hey1 | ventricular chamber development | [194,195,196] |
| Hey2 | ventricular chamber development | [194,195,196,197] |
| Coup-TFII | atrial development | [198] |
| eHand | ventricular development | [199,200,201,202] |
| dHand | ventricular development | [199,202] |
| Foxm1 | ventricular trabecular and compact layers development | [203] |
| Hop | ventricular trabecular and compact layers development | [204] |
| Klf13 | ventricular trabecular and compact layers development | [205] |
| Srf | ventricular trabecular and compact layers development | [206,207] |
| Shox2 | cardiac conduction development | [208,209,210,211,212,213] |
| Odd1 | atrial septation | [214] |
| Klf2 | atrioventricular septation | [215] |
| Sox9 | atrioventricular septation | [216] |
| Smad4 | atrioventricular septation | [217] |
| Tbx1 | outflow tract and aortic arch development | [218] |
| Foxc1 | aortic arch development | [219] |
| Foxc2 | aortic arch development | [219] |
| Prx1 | aortic arch development | [220] |
| Prx2 | aortic arch development | [220] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, D.; Garcia-Padilla, C.; Dominguez, J.N.; Lozano-Velasco, E.; Aranega, A. Cardiac Development: A Glimpse on Its Translational Contributions. Hearts 2021, 2, 87-118. https://0-doi-org.brum.beds.ac.uk/10.3390/hearts2010008
Franco D, Garcia-Padilla C, Dominguez JN, Lozano-Velasco E, Aranega A. Cardiac Development: A Glimpse on Its Translational Contributions. Hearts. 2021; 2(1):87-118. https://0-doi-org.brum.beds.ac.uk/10.3390/hearts2010008
Chicago/Turabian StyleFranco, Diego, Carlos Garcia-Padilla, Jorge N. Dominguez, Estefania Lozano-Velasco, and Amelia Aranega. 2021. "Cardiac Development: A Glimpse on Its Translational Contributions" Hearts 2, no. 1: 87-118. https://0-doi-org.brum.beds.ac.uk/10.3390/hearts2010008