Towards Tissue-Specific Stem Cell Therapy for the Intervertebral Disc: PPARδ Agonist Increases the Yield of Human Nucleus Pulposus Progenitor Cells in Expansion
Abstract
:1. Introduction
1.1. Nucleus Pulposus Cells and Their Progenitors
1.2. Tie2 and Self-Renewal
1.3. Mitophagy in NPC
1.4. Hypothesis and Aims
2. Materials and Methods
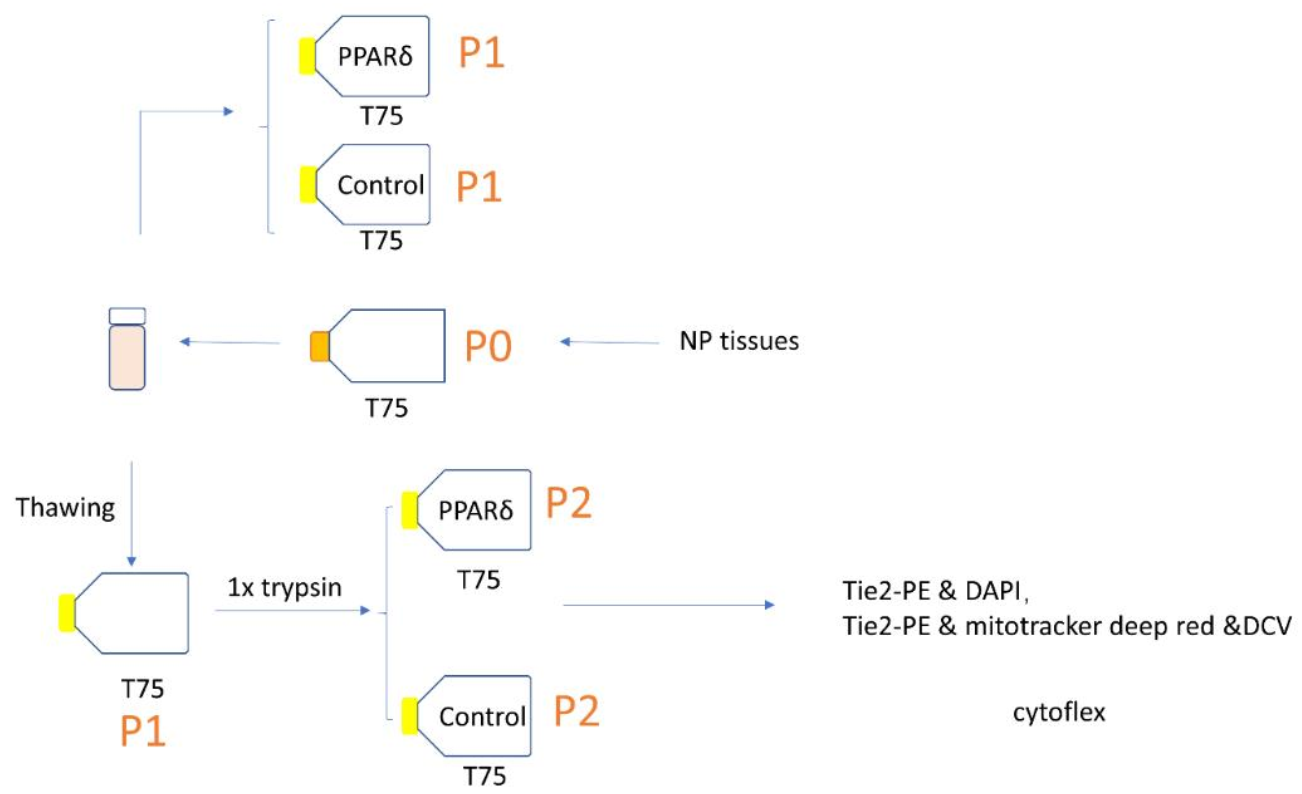
2.1. NPC Isolation and Culture
2.2. PPARδ Agonist Treatment
2.3. Visualisation of Cell’s Morphology
2.4. Flow Cytometry
2.5. qPCR
2.6. Population Doubling Level (PDL)
2.7. Statistics
3. Results
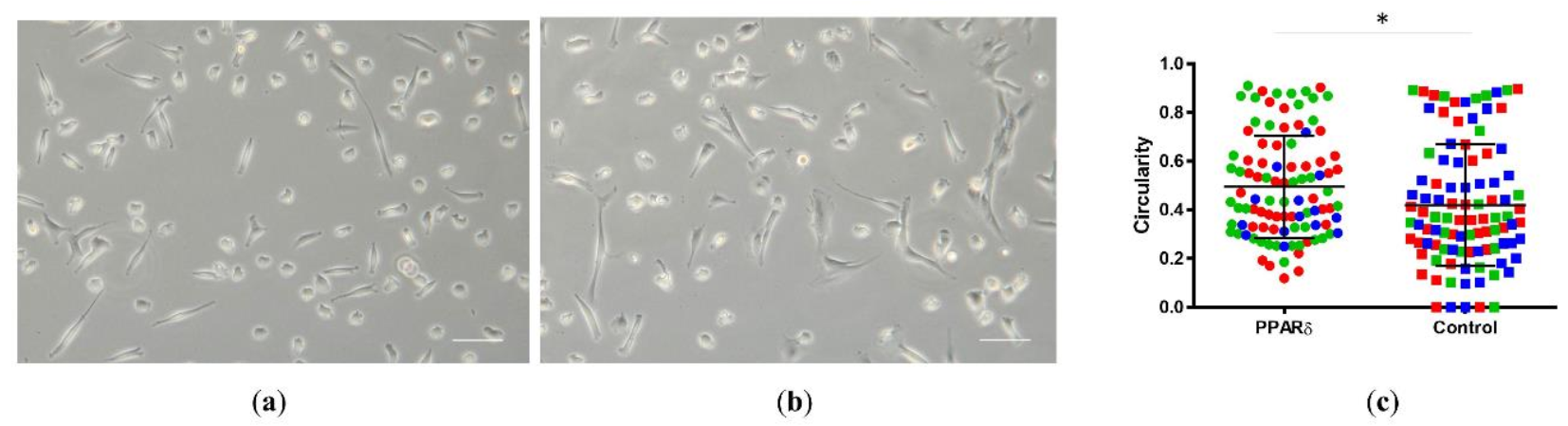
3.1. PPARδ Agonist Treatment Changed the Morphology of NPCs
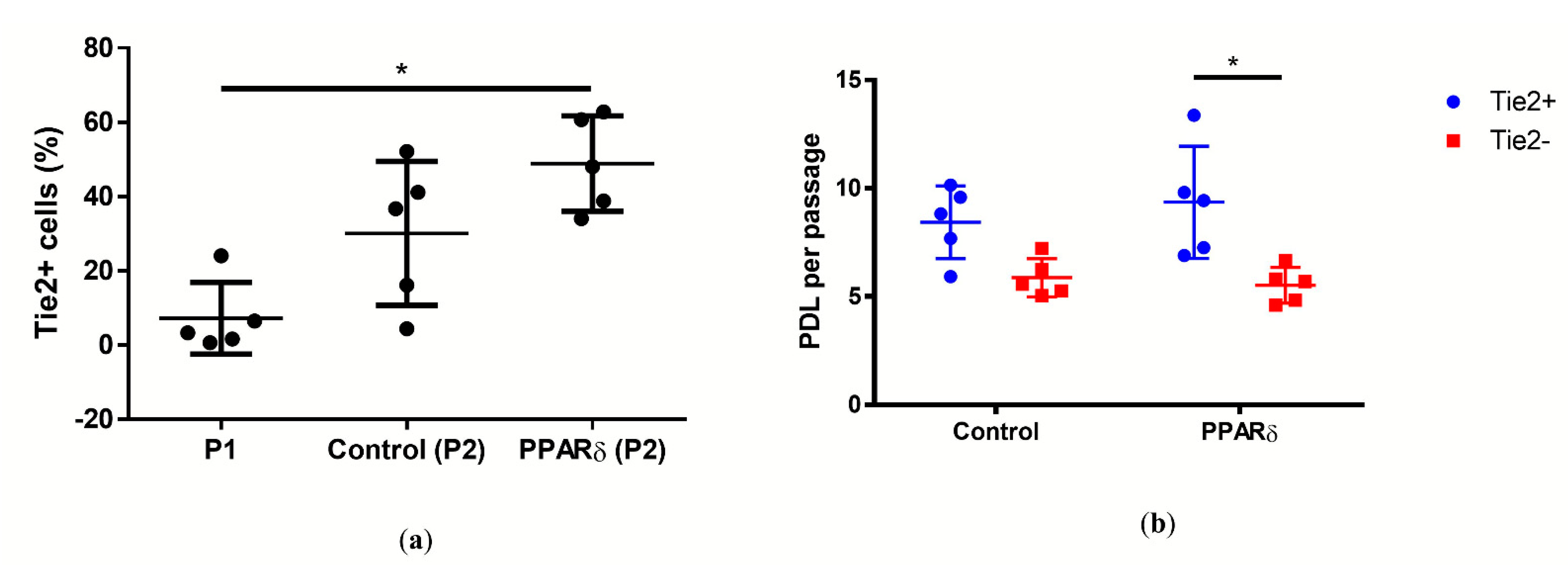
3.2. PPARδ Agonist Treatment Increased the Tie2+ NPC Yield of NPCs
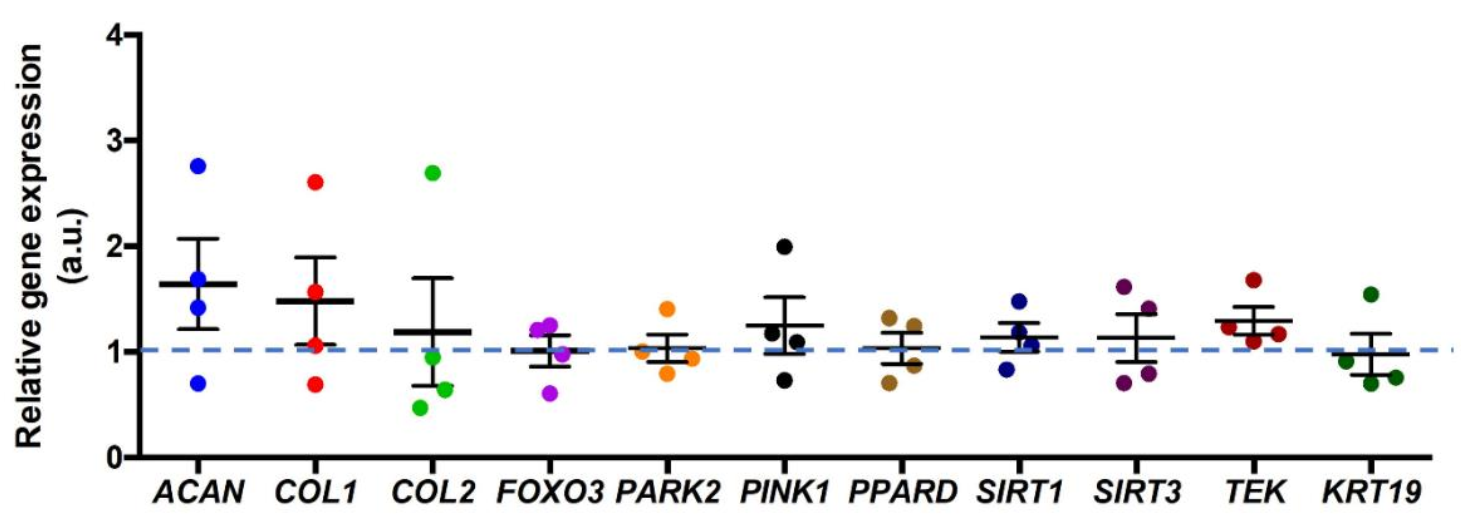
3.3. Correlation between mRNA Expression and PPARδ Agonist Treatment
3.4. Correlation between Mitochondrial Mass and PPARδ Agonist Treatment
4. Discussion
5. Conclusions
- The PPARδ agonist treatment significantly changed the cell’s morphology towards more rounded cells in 2D culture.
- The PPARδ agonist significantly accelerated cell proliferation in NPPCs, i.e., Tie2+ cells, compared to Tie2- cells, by a factor of ~7.
- The PPARδ agonist increased the Tie2+ yield after ~day eight of monolayer culture.
- The PPARδ agonist significantly decreased the mitochondria mass in NPPCs, i.e., Tie2+ cells, compared to Tie2- cells.
- The PPARδ agonist did not cause any significant changes in mitophagy-related relative gene expression, such as PARKIN and PINK1, nor in other ECM-related genes.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AF | Annulus fibrosus |
| bFGF | Fibroblast growth factor-basic |
| CEP | Cartilage endplates |
| CFU | Colony-forming unit |
| CFU-s | Spheroid colony-forming unit |
| CSA | Cyclosporine A |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DCV | Vybrant™ DyeCycle™ Violet |
| DMSO | Dimethyl sulfoxide |
| ECM | Extracellular matrix |
| EGF | Epidermal growth factor |
| EDTA | Ethylenediaminetetraacetic acid |
| K-W | Kruskal–Wallis signed-rank |
| LG-DMEM | Low-glucose Dulbecco’s Modified Eagle Medium |
| IVD | Intervertebral disc |
| IVDD | Intervertebral disc degeneration |
| MFI | Medians of fluorescence intensity |
| MSC | Mesenchymal stromal cell |
| NP | Nucleus pulposus |
| NPC | Nucleus pulposus cell |
| NPPC | Nucleus pulposus progenitor cell |
| P | Passage |
| PBS | Phosphate-buffered saline |
| P/S/G | Penicillin/Streptomycin/Glutamine |
| SD | Standard deviation |
References
- Frost, B.A.; Camarero-Espinosa, S.; Foster, E.J. Materials for the spine: Anatomy, problems, and solutions. Materials 2019, 12, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.; Evans, H.; Trivedi, J.; Menage, J. Histology and pathology of the human intervertebral disc. J. Bone Joint Surg. 2006, 88, 10–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, S.; Dahia, C.L. Defects in intervertebral disc and spine during development, degeneration, and pain: New research directions for disc regeneration and therapy. Dev. Biol. 2019, 8. [Google Scholar] [CrossRef]
- Williams, S.; Alkhatib, B.; Serra, R. Development of the axial skeleton and intervertebral disc. Curr. Top. Dev. Biol. 2019, 133, 49–90. [Google Scholar]
- Risbud, M.V.; Shapiro, I.M. Notochordal cells in the adult intervertebral disc: New perspective on an old question. Crit. Rev. Eukaryot. Gene Express. 2011, 21, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Colombier, P.; Clouet, J.; Boyer, C.; Ruel, M.; Bonin, G.; Lesoeur, J.; Moreau, A.; Fellah, B.-H.; Weiss, P.; Lescaudron, L.; et al. TGF-beta 1 and GDF5 act synergistically to drive the differentiation of human adipose stromal cells toward nucleus pulposus-like cells. Stem Cells 2016, 34, 653–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCann, M.R.; Seguin, C.A. Notochord cells in intervertebral disc development and degeneration. J. Dev. Biol. 2016, 4, 3. [Google Scholar] [CrossRef]
- Sakai, D.; Nakamura, Y.; Nakai, T.; Mishima, T.; Kato, S.; Grad, S.; Alini, M.; Risbud, M.V.; Chan, D.; Cheah, K.S. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat. Commun. 2012, 3, 1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tekari, A.; Chan, S.C.W.; Sakai, D.; Grad, S.; Gantenbein, B. Angiopoietin-1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res. Ther. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Frauchiger, D.A.; Tekari, A.; May, R.D.; Dzafo, E.; Chan, S.C.W.; Stoyanov, J.; Bertolo, A.; Zhang, X.; Guerrero, J.; Sakai, D.; et al. Fluorescence-activated cell sorting is more potent to fish intervertebral disk progenitor cells than magnetic and beads-based methods. Tissue Eng. Part C Methods 2019, 25, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Boyette, L.B.; Tuan, R.S.J.J. Adult stem cells and diseases of aging. J. Clin. Med. 2014, 3, 88–134. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.Y.; Moore, D.L. Neural stem cells: Developmental mechanisms and disease modeling. Cell Tissue Res. 2018, 371, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Williams, F.M.; Bansal, A.T.; Van Meurs, J.B.; Bell, J.T.; Meulenbelt, I.; Suri, P.; Rivadeneira, F.; Sambrook, P.N.; Hofman, A.; Bierma-Zeinstra, S. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: A meta-analysis of 4600 subjects. Ann. Rheumat. Dis. 2013, 72, 1141–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Näkki, A.; Battié, M.C.; Kaprio, J. Genetics of disc-related disorders: Current findings and lessons from other complex diseases. Eur. Spine J. 2014, 23, 354–363. [Google Scholar] [CrossRef]
- Madhu, V.; Boneski, P.K.; Silagi, E.; Qiu, Y.; Kurland, I.; Guntur, A.R.; Shapiro, I.M.; Risbud, M.V. Hypoxic regulation of mitochondrial metabolism and mitophagy in nucleus pulposus cells is dependent on HIF-1alpha-BNIP3 axis. J. Bone Min. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, J.; Häckel, S.; Croft, A.S.; Albers, C.E.; Gantenbein, B.J.J.S. The effects of 3D culture on the expansion and maintenance of nucleus pulposus progenitor cell multipotency. JOR Spine 2020, 1131. [Google Scholar] [CrossRef]
- Zhang, X.; Guerrero, J.; Croft, A.S.; Albers, C.E.; Häckel, S.; Gantenbein, B. Spheroid-like cultures for expanding angiopoietin receptor-1 (aka. Tie2) positive cells from the human intervertebral disc. Int. J. Mol. Sci. 2020, 21, 9423. [Google Scholar] [CrossRef]
- Teng, Y.D.; Liao, W.L.; Choi, H.; Konya, D.; Sabharwal, S.; Langer, R.; Sidman, R.L.; Snyder, E.Y.; Frontera, W.R. Physical activity-mediated functional recovery after spinal cord injury: Potential roles of neural stem cells. Regen Med. 2006, 1, 763–776. [Google Scholar] [CrossRef]
- Wangler, S.; Peroglio, M.; Menzel, U.; Benneker, L.M.; Haglund, L.; Sakai, D.; Alini, M.; Grad, S. Mesenchymal stem cell homing into intervertebral discs enhances the Tie2-positive progenitor cell population, prevents cell death, and induces a proliferative response. Spine 2019, 44, 1613–1622. [Google Scholar] [CrossRef]
- Ito, K.; Turcotte, R.; Cui, J.; Zimmerman, S.E.; Pinho, S.; Mizoguchi, T.; Arai, F.; Runnels, J.M.; Alt, C.; Teruya-Feldstein, J. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 2016, 354, 1156–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunmair, B.; Staniek, K.; Dörig, J.; Szöcs, Z.; Stadlbauer, K.; Marian, V.; Gras, F.; Anderwald, C.; Nohl, H.; Waldhäusl, W. Activation of PPAR-δ in isolated rat skeletal muscle switches fuel preference from glucose to fatty acids. Diabetologia 2006, 49, 2713–2722. [Google Scholar] [PubMed] [Green Version]
- Cairns, G.; Thumiah-Mootoo, M.; Burelle, Y.; Khacho, M. Mitophagy: A new player in stem cell biology. Biology 2020, 9, 481. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wu, Y.; Shi, H.; Wang, J.; Zheng, Z.; Chen, J.; Chen, X.; Zhang, Z.; Xu, D.; Wang, X.; et al. Melatonin ameliorates intervertebral disc degeneration via the potential mechanisms of mitophagy induction and apoptosis inhibition. J. Cell. Mol. Med. 2019, 23, 2136–2148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Shen, J.; Chen, Y.; Liu, H.; Zhou, H.; Bai, Z.; Hu, Z.; Guo, X. PINK1 protects against oxidative stress induced senescence of human nucleus pulposus cells via regulating mitophagy. Biochem. Biophys. Res. Commun. 2018, 504, 406–414. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, T.; Chen, J.; Shao, Z.; Wang, K.; Yan, Y.; Wu, C.; Lin, J.; Wan, H.; Gao, W.; et al. Parkin-mediated mitophagy as a potential therapeutic target for intervertebral disc degeneration. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Castro, E.; Cunningham, C.J.; Miller, J.; Cain, S.A.; Allan, S.M.; Pinteaux, E. Generation of Human Mesenchymal Stem Cell 3D Spheroids Using Low-binding Plates. Bio Protoc. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasband, W.S. ImageJ, U.S. National Institutes of Health, Bethesda, MD, USA. Available online: http://imagej.nih.gov/ij/ (accessed on 15 February 2021).
- Barcellona, M.N.; Speer, J.E.; Fearing, B.V.; Jing, L.; Pathak, A.; Gupta, M.C.; Buchowski, J.M.; Kelly, M.; Setton, L.A. Control of adhesive ligand density for modulation of nucleus pulposus cell phenotype. Biomaterials 2020, 250, 120057. [Google Scholar] [CrossRef] [PubMed]
- Gantenbein-Ritter, B.; Benneker, L.M.; Alini, M.; Grad, S. Differential response of human bone marrow stromal cells to either TGF-β(1) or rhGDF-5. Eur. Spine J. 2011, 20, 962–971. [Google Scholar] [CrossRef] [Green Version]
- Mauro-Lizcano, M.; Esteban-Martínez, L.; Seco, E.; Serrano-Puebla, A.; Garcia-Ledo, L.; Figueiredo-Pereira, C.; Vieira, H.L.; Boya, P. New method to assess mitophagy flux by flow cytometry. Autophagy 2015, 11, 833–843. [Google Scholar] [CrossRef]
- Farnie, G.; Sotgia, F.; Lisanti, M.P. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget 2015, 6, 30472–30486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannini, N.; Girotra, M.; Naveiras, O.; Nikitin, G.; Campos, V.; Giger, S.; Roch, A.; Auwerx, J.; Lutolf, M.P. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat. Commun. 2016, 7, 13125. [Google Scholar] [CrossRef] [PubMed]
- Takihara, Y.; Nakamura-Ishizu, A.; Tan, D.Q.; Fukuda, M.; Matsumura, T.; Endoh, M.; Arima, Y.; Chin, D.W.L.; Umemoto, T.; Hashimoto, M.; et al. High mitochondrial mass is associated with reconstitution capacity and quiescence of hematopoietic stem cells. Blood Adv. 2019, 3, 2323–2327. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Peng, Y.; Li, Z.; Chen, S.; Deng, X.; Shao, Z.; Ma, K. Compression-induced senescence of nucleus pulposus cells by promoting mitophagy activation via the PINK1/PARKIN pathway. J. Cell. Mol. Med. 2020, 24, 5850–5864. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No. | Gender | Age | Pfirrmann Grade * | Location |
|---|---|---|---|---|
| 1 | female | 37 | 1 | T12/L1 |
| 2 | female | 39 | 2 | L2/L3 |
| 3 | male | 40 | 2 | L2/L3 |
| 4 | male | 19 | 1 | T12/L1 |
| 5 | male | 28 | 1 | L4/L5 |
| 6 | male | 38 | 2 | L4/5 |
| 7 | male | 23 | 1-2 | T11/12 |
| 8 | male | 40 | 2 | T11/T1 |
| Name. | Description | Primer Forward | Primer Reverse |
|---|---|---|---|
| 18S | Ribosomal 18s RNA gene | CGA TGC GGC GGC GTT ATT C | TCT GTC AAT CCT GTC CGT GTC C |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | ATC TTC CAG GAG CGA GAT | GGA GGC ATT GCT GAT GAT |
| ACAN | Aggrecan core protein | CAT CAC TGC AGC TGT CAC | AGC AGC ACT ACC TCC TTC |
| COL1 | Collagen type 1 | GTG GCA GTG ATG GAA GTG | CAC CAG TAA GGC CGT TTG |
| COL2 | Collagen type 2 | AGC AGC AAG AGC AAG GAG AA | GTA GGA AGG TCA TCT GGA |
| KRT19 | Keratin 19 | TGT GTC CTC GTC CTC CTC | GCG GAT CTT CAC CTC TAG C |
| TEK | TEK receptor tyrosine kinase | TTA GCC AGC TTA GTT CTC TGT GG | AGC ATC AGA TAC AAG AGG TAG GG |
| PARK2 | Parkin RBR E3 Ubiquitin Protein Ligase | GTC TTT GTC AGG TTC AAC TCC A | GAA AAT CAC ACG CAA CTG GTC |
| PINK1 | Phosphatase and tensin homolog deleted on chromosome ten (PTEN)-induced putative kinase 1 | CTCCAGCGAAGCCATCTT | TCTGTAAGTGACTGCTCCATAC |
| PPARD | Peroxisome proliferator-activated receptor delta | CAG GGC TGA CTG CAA ACG A | CTG CCA CAA TGT CTC GAT GTC |
| SIRT1 | Silent mating type information regulation 2 homolog 1 | TAGCCTTGTCAGATAAGGAAGGA | ACAGCTTCACAGTCAACTTTGT |
| SIRT3 | Silent mating type information regulation 2 homolog 3 | CAG CAA CCT CCA GCA GTA | CGT GTA GAG CCG CAG AAG |
| FOXO3 | Forkhead Box O3 | CGG ACA AAC GGC TCA CTC T | GGA CCC GCA TGA ATC GAC TAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Guerrero, J.; Croft, A.S.; Oswald, K.A.C.; Albers, C.E.; Häckel, S.; Gantenbein, B. Towards Tissue-Specific Stem Cell Therapy for the Intervertebral Disc: PPARδ Agonist Increases the Yield of Human Nucleus Pulposus Progenitor Cells in Expansion. Surgeries 2021, 2, 92-104. https://0-doi-org.brum.beds.ac.uk/10.3390/surgeries2010008
Zhang X, Guerrero J, Croft AS, Oswald KAC, Albers CE, Häckel S, Gantenbein B. Towards Tissue-Specific Stem Cell Therapy for the Intervertebral Disc: PPARδ Agonist Increases the Yield of Human Nucleus Pulposus Progenitor Cells in Expansion. Surgeries. 2021; 2(1):92-104. https://0-doi-org.brum.beds.ac.uk/10.3390/surgeries2010008
Chicago/Turabian StyleZhang, Xingshuo, Julien Guerrero, Andreas S. Croft, Katharina A.C. Oswald, Christoph E. Albers, Sonja Häckel, and Benjamin Gantenbein. 2021. "Towards Tissue-Specific Stem Cell Therapy for the Intervertebral Disc: PPARδ Agonist Increases the Yield of Human Nucleus Pulposus Progenitor Cells in Expansion" Surgeries 2, no. 1: 92-104. https://0-doi-org.brum.beds.ac.uk/10.3390/surgeries2010008







