1. Introduction
Alzheimer’s disease (AD) is one of the diseases that is among the trends of epidemics for the coming years, due to the increase of life expectancy of the population, since the incidence of this disease grows exponentially with age [
1]. The AD corresponds to the most common form of dementia, affecting more than 46 million people worldwide, with expectation of going up to 130 million cases by 2050 [
2]. Despite the high worldwide incidence, only 1 in 4 people with AD are diagnosed [
3]. Statistics from 2011 revealed that approximately 36 million people had the disease in the world, but about 28 million did not receive the proper diagnostic [
4]. These data highlight one of the main current concerns related to the disease: its differential and early diagnostic.
The symptoms of early-stage AD may be easily confused with those of other pathologies, since other diseases also have one of the main characteristics of memory lapses [
5,
6]. Thereunto, in 2011, the National Institute on Aging-Alzheimer’s Association (NIA-AA) suggested the use of biomarkers as a new criterion for the identification of AD in the asymptomatic phase [
7]. The use of qualifying and quantifying biomarkers of biochemical changes in the brain decreases probable diagnostic errors and provides access to appropriate treatments even before clinical signs appear, minimizing the impacts caused by the disease, especially with regard to the loss of cognitive functions [
8,
9].
Tau is a protein with great preclinical relevance, because it is directly related to neuronal death, which is the main clinical sign of AD. In its normal function (i.e., physiological function), this protein is responsible for the stabilization of the microtubules, allowing the passage of current in the axons [
10]. The function of tau is compromised when there is a hyperphosphorylation, reducing the attachment to the microtubules and forming paired helical filament (PHF) agglomerates in the extracellular spaces, known as neurofibrillary tangles [
11]. Neurofibrillary tangles spread in the brain as AD progresses and cause synaptic loss and gradual neuronal death in neurons [
12].
PHF-tau can be considered a promising pre-symptomatic biomarker of AD, since aggregates of this protein can be found in the brain years before the symptoms appear [
11]. Nanoscale sensors have been suggested for the detection of biological markers associated with various diseases at the cellular and molecular levels [
13]. In addition, the use of nanomaterials in the implementation of biosensors increases the speed, sensitivity and specificity of detection by the biological molecule of interest [
14].
Carbon-based nanomaterials are being widely used in the sensing of different diseases [
15,
16]. Particularly, the fullerol is an interesting and versatile nanomaterial derived belonging to the fullerene family. This structure can be efficiently functionalized (i.e., oxidized) with hydroxyl groups (-OH), thus making the molecule of the fullerene more hydrophilic and improving its bioavailability-based pharmacokinetics properties [
17]. Fullerene is a promising nanocarrier with respect to the passage of molecules through the blood–brain barrier (BBB), besides having excellent antioxidant and neuroprotective activity that was experimentally validated by in vitro experiments, improving, even, the neuronal function in AD [
18,
19,
20]. At the same time, a series of fluorescent radiopharmaceuticals have been studied experimentally for the diagnostic of several pathophysiological processes in the brain [
21,
22]. These studies have demonstrated that these substances have an affinity for the PHF-tau protein and can be used as receptors of this protein in biosensors [
22].
In this context, computational approaches could be efficiently applied for fast, cheap, and early detection of AD as an alternative strategy previous to perform experimental studies, allowing a better understanding of the molecular mechanisms of diagnostic recognition by using computational animal free-testing methodologies [
23,
24,
25]. During studies with nanostructures, the computational simulation assists in the understanding of physical and chemical phenomena occurring on the nanometric scale [
2].
In this context, the main objective of the present study was to implement and obtain a prospective and computationally modeled new nanomarker formed by functionalized fullerol with nine radiopharmaceutical ligands, with the potential ability of detecting early AD from the PHF-tau biomarker protein, through a combined computational approach like ab initio DFT and molecular docking simulation. To this end, the free energy of binding between the diagnostic systems (i.e., the fullerol with each radiopharmaceutical ligands) with the biomolecule (tau protein receptor) will be predicted with the corresponding molecular distances. Next, the mechanism of interaction for the best fit docking pose configuration will be studied through ab initio DFT simulation.
2. Materials and Methods
Firstly, the fullerol (C
60(OH)
24) was used as receptor for adding the fluorescent radiopharmaceuticals as
18F-FDDNP,
18F-T807,
18F-T808,
18F-THK523,
18F-THK5105,
18F-THK5117,
18F-THK5351,
11C-
N-methyl Lansoprazole and
11C-PBB3. The radiopharmaceutical molecules were obtained through the PubChem database [
26]. The immobilization of the radiopharmaceuticals to fullerol was performed through covalent bonds from the removal of water molecules from the system. Different configurations of functionalization of fullerol were analyzed with each radiopharmaceutical (nine in total). This analysis was done through ab initio calculations based on the density functional theory (DFT) using Spanish initiative for electronic simulations with thousands of atoms (SIESTA) computer code [
27]. The exchange-correlation term in the SIESTA code was reproduced by using the local density approximation (LDA) [
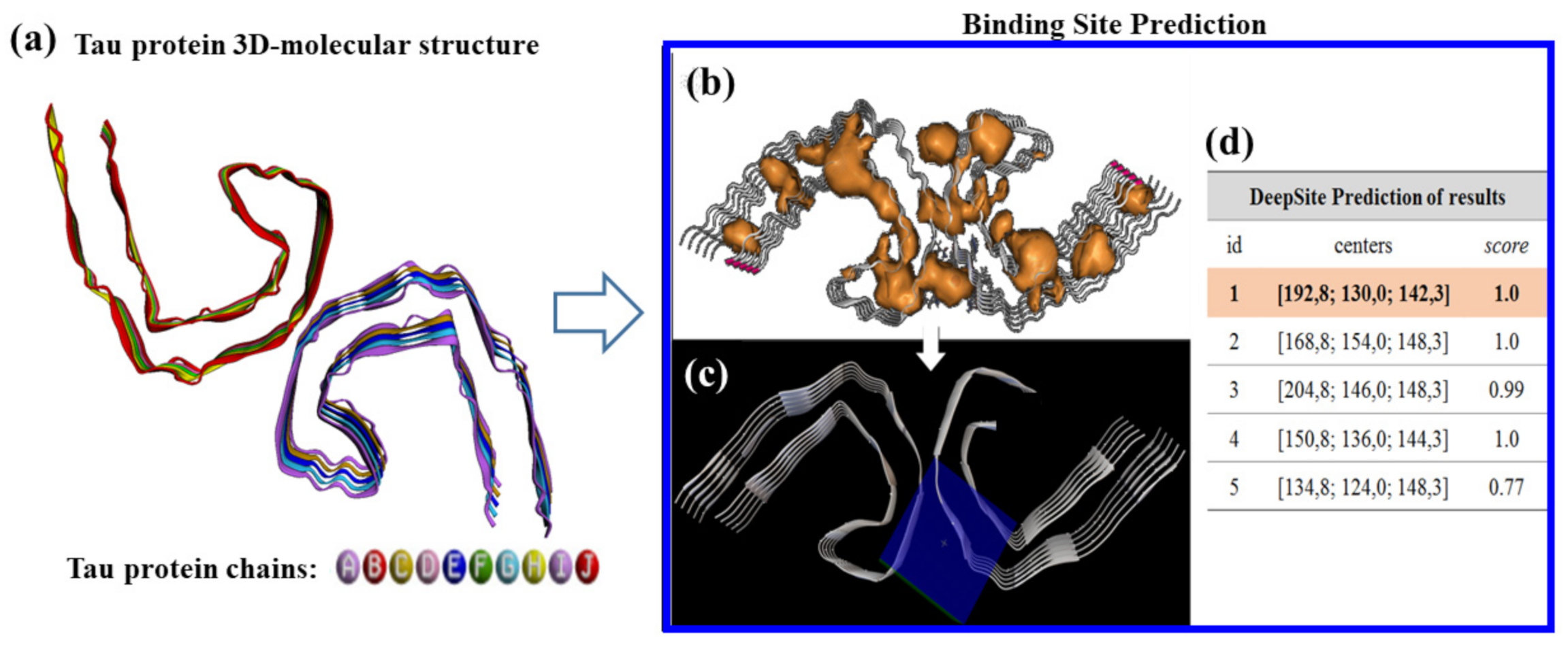
28]. The most stable fullerol-radiopharmaceutical systems were used as input ligands to address a rigid molecular docking study. Before the rigid docking execution, the potential tau protein binding-sites were predicted by using DeepSite [
29]. This task was carried out here by delimiting the van der Waals surface area of the tau-protein binding residues, where the binds of the ligands (i.e., fullerol-radiopharmaceutical systems) take place. The DeepSite method (freely available at
www.playmolecule.org , accessed on 15 March 2021) [
29] is mainly based on a 3D-deep convolutional neural networks approach (DCNNs) which efficiently predict the resulting tau-protein docking box of simulation. For this instance, the Cartesian coordinates generated from the DeepSite volumetric maps of the tau-protein binding sites were used to setup a rigid-docking box simulation in the best-ranked binding site of tau protein, considering the DeepSite maximum score and maximum volume of the selected cavity, which is well-known to have the highest druggability properties or ability to binds small ligands like fullerol-radiopharmaceutical systems. Them for the tau-protein, the details of the box-simulation are: grid box-size Cartesian coordinates of (
X = 27 Å,
Y = 27 Å,
Z = 27 Å) with volume V = 827.02 Å
3, and grid box-center Cartesian coordinates centered at
X = 192.8 Å,
Y = 130 Å,
Z = 142.3 Å. Afterward, all the molecular docking runs were performed using the AutoDock Vina package, with a Vina scoring function implemented by Trott and Olson. [
30,
31,
32]. Molecular docking approach is based on the prediction of best crystallographic binding conformation and the determination of the free energy of binding (FEB, kcal/mol) which quantify the affinity between the protein-binding complexes. To this end, the macromolecule (tau-protein), was obtained from the Protein Data Bank (PDB) website [
30], as PDB ID: 5O3L, resolution 3.4 Å was pretreated through of the AutoDock and PyMOL softwares for the removal of crystallographic water molecules and ions.
The best crystallographic binding configuration was check for the best fitting structure (fullerol-radiopharmaceutical systems), and based on two criteria: (i) most negative affinity or FEB value from the obtained docking complexes; and (ii) the root-mean- square-deviation (R.M.S.D) for the atoms positions for the obtained docking complexes with an optimal value of R.M.S.D < 2 Å for all the simulation experiments [
31]. In order to provide more reliable and accurate results, the AutoDock exhaustiveness parameter equal to 50 (i.e., representing 50 fullerol-radiopharmaceutical docking-pose configurations in the modeled rigid-pocket of tau-protein) was established [
32].
The specific regions of the docking interactions, as well as the type of interactions, were verified through the LigPlot program [
33]. This program evaluates the performance of non-covalent interactions between a protein and the ligand through a 2D diagram that shows the shape of interaction that occurred in the coupling (hydrophobic, hydrogen bonding, and electrostatics interactions) in the formed docking complexes as fullerol-radiopharmaceutical plus tau-protein. The resulting images of the atoms involved in the interactions of the radio-ligands and the atoms of the interacting amino-acid residues of tau-protein, as well as the corresponding values of the inter-atomic distances, were depicted by using PyMOL.
The interface of the post-docking system was studied through ab initio calculations using the SIESTA code.
Then, the electronic and structural properties between the best diagnostic system composed by the fullerol-radiopharmaceutical ligand (i.e., more stable configuration obtained by DFT approach), and the most critical amino acid residue from tau protein (i.e., identified with the shortest interaction distance) were evaluated according to the binding energy of the system calculated as show the Equation (1):
where
is the total energy (eV) of the interface of the amino acid + ligand system,
energy from isolated amino acid and
is the isolated binding energy.
Furthermore, the difference between the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) (ΔHL) was also calculated. The isosurface used for analyzed the charges concentrations in the HOMO and LUMO was 0.001 e-/Bohr
3 [
27]. Next, the molecular structures were depicted using the VMD program. [
34].
4. Discussion
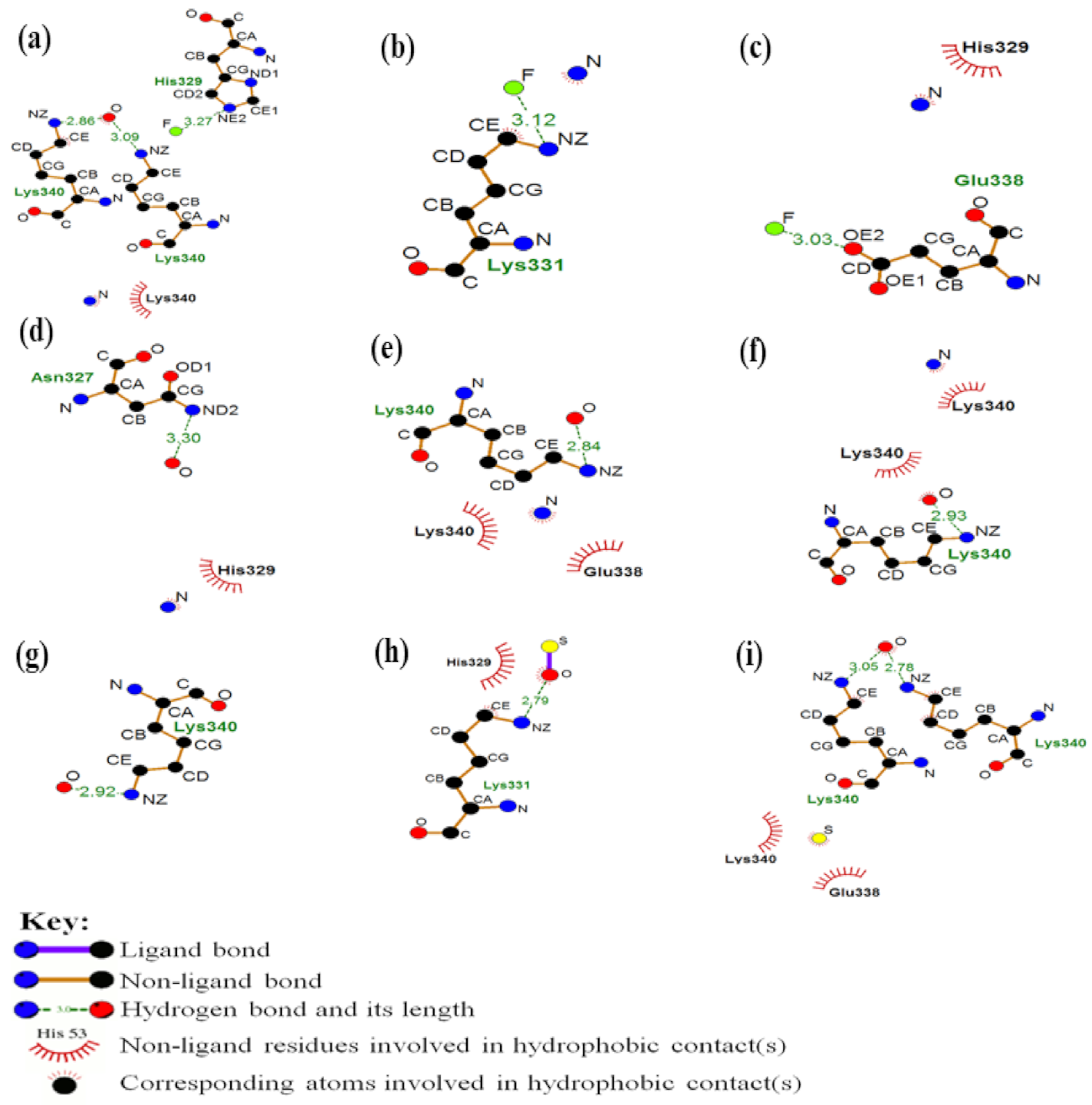
As observed in
Figure 3, in system (a) the atoms of the ligand involved interactions of fluorine and oxygen atoms. For this instance, the fluorine interacted with the histidine residue of the tau protein through a hydrogen bond, with a distance of 3.3 Å. Yet, the oxygen interacted through hydrogen bonds with the lysine residues, with distances of 2.9 and 3.1 Å. In addition, was observed a hydrophobic interaction between the nitrogen atom and the lysine residue of the protein.
For system (b), the region where direct interaction occurs involves the fluorine atom and the lysine residue of the protein, interrelating from a hydrogen bond. The distance from this interaction was 3.1Å. Besides to hydrogen bonding, a hydrophobic interaction occurred between the nitrogen atom of the ligand and the carbon atom of the protein residue.
In the arrangement of the system (c), it occurs a hydrogen bond of 3.0 Å of distance between the fluorine and the glutamate residue of the protein. In addition, a hydrophobic contact occurred between the nitrogen of the ligand and the histidine residue of the macromolecule.
In the complex (d), there was a hydrogen bond of 3.3 Å between the oxygen of the ligand and the nitrogen atom of the asparagine residue. There was also a hydrophobic contact between the ligand nitrogen and the histidine residue of the protein.
For best-ranked docking system (i.e., (e): PHF—
18F-THK5105 + fullerol), we show that the oxygen atom of the ligand interacted through a hydrogen bond of 2.8 Å with the lysine residue. There were also two hydrophobic contacts between the nitrogen atom of the ligand and the lysine and glutamate residues of the macromolecule. Details are depicted in the
Figure 4 showing three-dimensional image representation of the whole docking system as PHF—
18F-THK5105 + fullerol represented in (e).
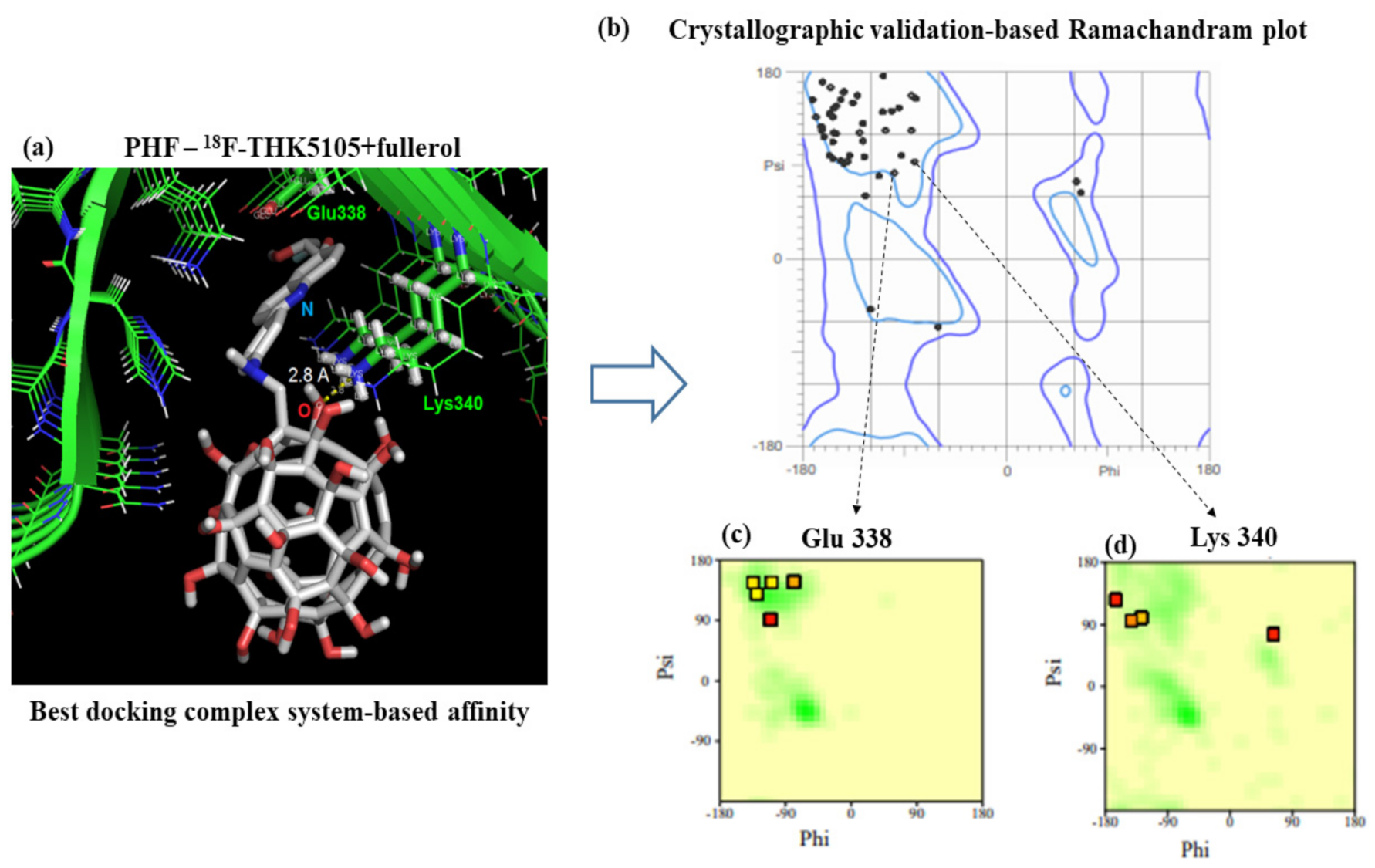
As shown in
Figure 4, the crystallographic validation results show that the 100% of the residues composing the tau protein can be categorized like conformationally favored residues with total absence of restricted flexibility. This fact presents a paramount relevance because it prevents the presence of potential false positives and results in the modeling of docking interactions, as well as ensuring the quality of the results from the conformational point of view for the identified interactions with the target residues. This allows the obtainment of a more proper evaluation on the conformational integrity of the target-residues potentially involved in the molecular recognition process toward biomedical detection purposes.
The remaining systems, such as (f), there was a hydrogen bond of 2.9 Å between the oxygen of the ligand and the nitrogen of the lysine residue. Furthermore, two hydrophobic contacts occurred, one between the same oxygen atom of the ligand and the lysine residue, and another between the nitrogen atoms of the ligand the lysine residue. In the docking complex (g), the main interaction of the system occurred through a hydrogen bond of 2.9 Å between the oxygen of the ligand and the nitrogen of the residue lysine.
In the arrangement (h) a hydrogen bond of 2.8 Å occurred between the oxygen of the ligand and the nitrogen of the lysine residue. In addition, there was a hydrophobic contact between the histidine residue and the same oxygen atom of the ligand.
Finally, in the system (i) it can be observed that two hydrogen bonds occurred with the oxygen atom of the ligand and lysine residues, with distances of 2.8 and 3.1 Å. In addition, there were also two hydrophobic contacts with the sulfur atom of the ligand and the residues lysine and glutamate.
In the analysis, the interactions that involved the systems were those of hydrophobic contact and hydrogen bonding. Hydrophobic interactions occur between non-polar groups, and although they are considered relatively weak interactions when compared to the others, these interactions are of great relevance in the complexation process between the protein and the ligand [
35,
36]. In addition, all distances were less than 7 Å, indicating a higher likelihood of interaction in terms of affinity and less chance of false contacts [
31].
The best complementarity feature according to docking criteria was obtained for the PHF—
18F-THK5105 + fullerol ligand system. As observed in
Figure 4, system (e), the interface of this docking system is composed of the lysine residue (Lys340) interacting through a hydrogen bond of 2.8 Å with the oxygen atom of the PHF—
18F-THK5105 + fullerol ligand system. Based on this, the structural and electronic properties of the interaction interface of this complex were studied, as shown in
Figure 5.
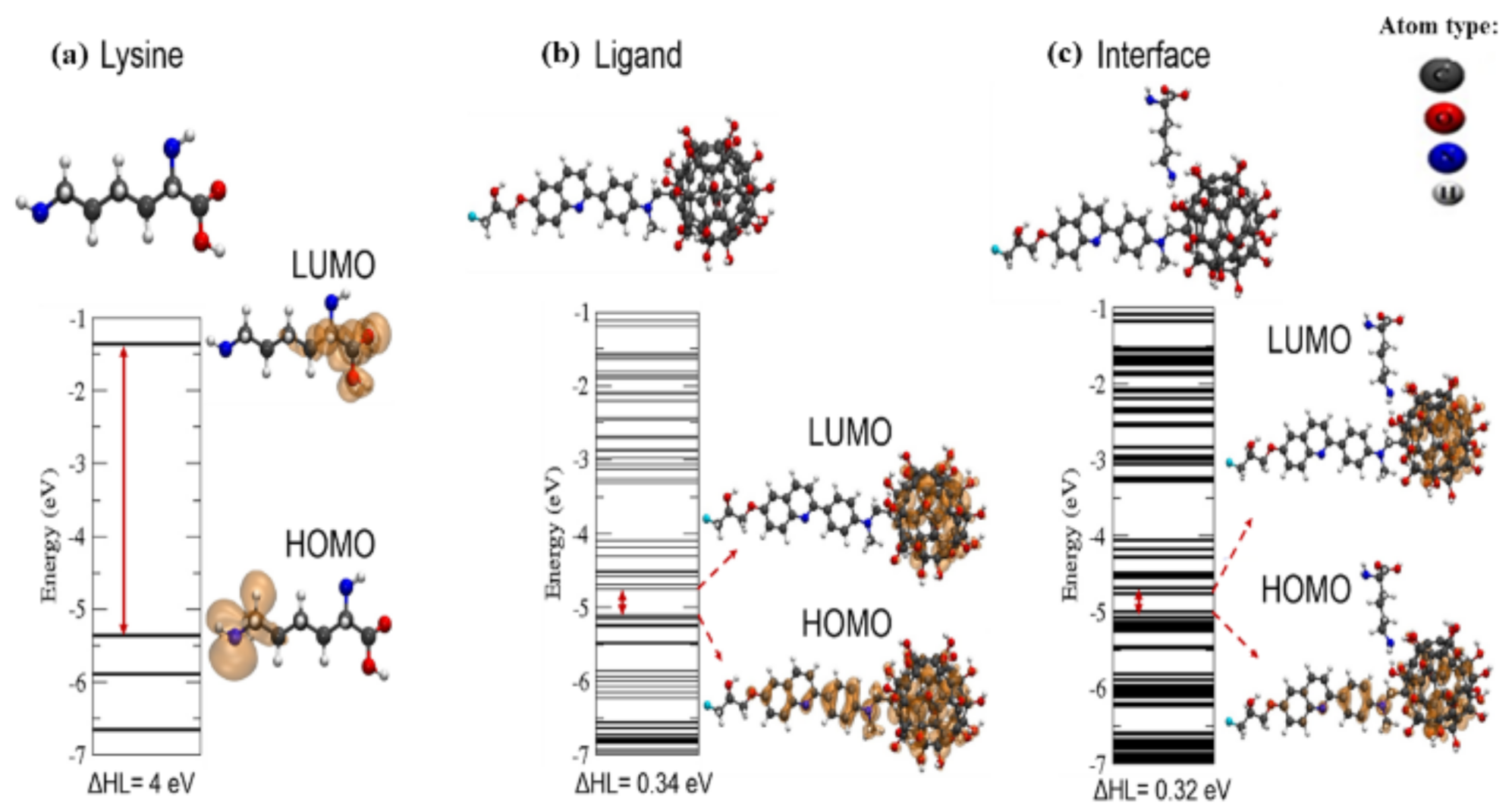
As shown above in
Figure 5, the electron density charges from LUMO and HOMO levels of the isolated structures (tau amino acid Lys34 and the PHF—
18F-THK5105 + fullerol ligand) fit well with the docking results obtained for the interface of interaction (see
Figure 4a). For this instance, the electron density charges were mainly concentrated over the ligand (PHF—
18F-THK5105 + fullerol system) which is a relevant theoretical evidence for potential biomedical application toward the rational design of new diagnostic nanomarker. In addition, considering the results obtained in the
Table 2 we can observe that there were no significant changes between the final distance and the initial distance of the system, proving the results of the docking. The negative interaction energy (−0.9 eV) clearly indicates attraction between the structures (i.e., Lys340 tau-protein and ligand PHF—
18F-THK5105 + fullerol). Lastly, the obtained theoretical results strongly suggest that the docking system is formed through a physical adsorption regime, which is an attribute of great relevance to be considered in the in the development of molecular detection system of high selectivity and specificity.
In addition, it is important to highlight that, theoretically, from the therapeutic point of view, for the main advantages of using nanotechnology-based diagnosis markers over traditional methods of biomarker diagnosis we can refer to the following: (i) the possibility of significantly reducing the dose to be administered during the diagnostic procedures compared with conventional diagnostic procedures with better cost/benefit relationships, (ii) significant increase of the bioavailability, as well as, optimal pharmacokinetics atributes namely: absorption, distribution, metabolism and elimination (ADME), (iii) overcome the blood–brain barrier (BBB), which significantly limits the entry of micro-scale diagnostic compounds compared with the nano-scale-based drugs allowing the more easy access to specific areas of the nervous tissue, in early stages of the Alzheimer’s disease, (iv) in consonance, the use of nanomarkers-based fullerol has theoretically an optimal balance between the lipophilic/hydrophilic properties over the conventional diagnostic systems based on its expected high lipid/water partition coefficient (Log P) that could favor the easy passage across the BBB, easy metabolism and elimination by the biochemical mechanisms of biotransformation (phase II) in the brain.
Lastly, the use of nanotechnology-based markers over traditional methods of diagnosis could significantly improve new therapeutic strategies of precision nanomedicine.
5. Conclusions
The nanosystem to be used as nanobiomarker was computationally modeled and the binding interaction (affinity) of the nanostructured ligands with the PHF-tau protein was evaluated through a combination of molecular docking and ab initio DFT simulation. The in silico results suggest that, all the nano-ligands studied present good affinity with optimal inter-atomic distances of interaction, suggesting high tau protein specificity and selectivity for AD detection purposes. The evaluated nanomarkers ligands demonstrated a predominance of hydrophobic and hydrogen bond contacts with the target tau protein (binding site ID 1); which are considered as relevant for molecular recognition purposes for potential biomedical applications.
The best docking complex based on the binding affinity by PHF-tau protein was the F-THK5105 plus fullerol, with a binding energy value of −7.0 kcal/mol and R.M.S.D value of 1.491 Å. In this case, the best evaluated interface of interaction of the nanomarker system (i.e., Lys340 interacting with atoms from the PHF—18F-THK5105 + fullerol) shows no significant changes to its electronic and structural properties, pointing a physical adsorption regime of interactions. These results fit with the obtained results from the molecular docking approach, and strongly suggest that the nanosystem presents promising properties and optimal features toward sensing and molecular recognition applications.
Thus, all the arrangements studied present characteristics of complementarity and could be considered as promising for the development of a therapeutically relevant nanomarker for the early detection of Alzheimer’s disease. As a future perspective, we will evaluate the molecular dynamics based on binding properties of the modeled nanostructures interacting with the cell wall from neurons to the inside of the cytoplasm in the context of in vitro and in vivo experiments.
Lastly, the obtained results open new avenues in the computational modeling in neuroscience by combining molecular docking recognition and DFT approaches for early detection of Alzheimer’s diseases.