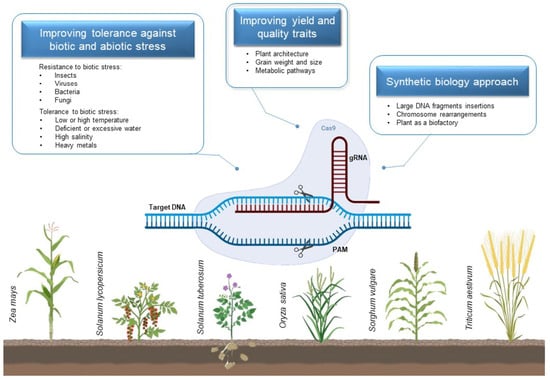
CRISPR towards a Sustainable Agriculture
Definition
:1. Introduction
1.1. CRISPR/Cas System
1.2. CRISPR Variants, Orthologs and Engineered Systems
1.3. New Perspectives for the Use of CRISPR/Cas System
2. Applications
| Application | Plant Species | Target Genes | Resulting Traits | References |
|---|---|---|---|---|
| Genetic Variability | Rice Pea Tomato Wheat | Zep1 RECQ4 ZIP4-B2 | Enhanced genetic recombination frequency Increased crossover frequency Enhanced recombination between homeologous chromosomes | [50] |
| [51] | ||||
| [52] | ||||
| Stress tolerance | Rice Maize Wheat Soybean Potato Tomato | TIFY1b ERF922 eIF4G Als2 ARGOS8 MLO Qsd1 F3H1, F3H2, FNSII-1 ALS RXLR effector gene Avr 4/6 Mlo1 ACET1a, ACET1b MAPK3 | Improved adaptation to low temperature Improved resistance to rice blast Improved resistance to rice tungro spherical virus Chlorsulfuron-resistant maize Improved resistance to drought Enhanced resistance to powdery mildew Longer dormancy Increased isoflavone content and resistance to soybean mosaic virus Development of glyphosate-resistant soybean Enhanced tolerance to Phytophthora infestans Reduced powdery mildew susceptibility Increased resistance to Botrytis cinerea Enhanced tolerance to heat stress | [53] |
| [54] | ||||
| [55] | ||||
| [56] | ||||
| [57] | ||||
| [58] | ||||
| [59] | ||||
| [60] | ||||
| [61] | ||||
| [62] | ||||
| [31] | ||||
| [63] | ||||
| [64] | ||||
| Yield | Rice Maize Wheat Soybean Tomato | GN1a, DEP1, GS3 GW2, GW5, TGW6 PYL1, PYL4, PYL6 CLE GW2 GASR7 GW7 FT2a, FT5a fas, lc | Enhanced grain number and size, dense erect panicles Enhanced grain weight Improved growth and productivity Enhanced kernel number Enhanced grain size and weight Enhanced grain size Enhanced grain size and weight Increased numbers of pods and seeds Enhanced fruit size | [65] |
| [66] | ||||
| [67] | ||||
| [68] | ||||
| [69] | ||||
| [70] | ||||
| [71] | ||||
| [72] | ||||
| [73] | ||||
| Quality | Rice Maize Wheat Potato Sweet potato Tomato Barley Rapeseed | CrtI, PSY GAD3 SBEIIb, SBEI GBSS GBSS IPK1 SBEIIa α-gliadin genes CM3, CM16 ASN2 SBEI GBSS SBEII SGR1, LCY-E, Blc, LCY-B1, LCY-B2 GAD2, GAD3 GABA-Ts, SSADH GBSS D-hordein ITPK | High β-carotene content High GABA content High amylose content Low amylose content Low amylose content Low phytic acid content High amylose content Low gluten content Reduced amount of potential allergens Reduced free asparagine High amylose content Low amylose content High amylose content High lycopene accumulation High GABA content High GABA content Low amylose content Reduced prolamine content and increased glutenin content Low phytic acid content | [33] |
| [74] | ||||
| [75] | ||||
| [76] | ||||
| [77] | ||||
| [78] | ||||
| [79] | ||||
| [80] | ||||
| [81] | ||||
| [82] | ||||
| [83] | ||||
| [84] | ||||
| [84] | ||||
| [85] | ||||
| [86] | ||||
| [87] | ||||
| [88] | ||||
| [89] | ||||
| [90] | ||||
| Synthetic Biology | Rice Potato Tomato Barley Arabidopsis Switchgrass Rapeseed Salvia Tobacco | CrtI, PSY >GBSS PSY1 CRTISO COMT-1 Chromosome 1, Chromosome 2 4CL FAD2 CPS1 NtAn1 | Insertion of large DNA fragments Low amylose content Inter-homologous somatic recombination Inter-homologous somatic recombination Increased bioethanol concentration of the mutant biomass Reciprocal translocation Used as lignocellulosic feedstock for bioenergy Increased content of oleic acid Customization of secondary metabolite profiles Increased seed lipid accumulation for biodiesel production | [33] |
| [91] | ||||
| [92] | ||||
| [93] | ||||
| [94] | ||||
| [95] | ||||
| [96] | ||||
| [97] | ||||
| [98] | ||||
| [99] |
2.1. Broadening Genetic Variability
2.1.1. Enhancing Stress Tolerance
2.1.2. High Yield
2.1.3. Quality Improvement
2.2. Synthetic Biology
3. Legislation Limits
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Entry Link on the Encyclopedia Platform
References
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 80, 346. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Urrea Castellanos, R.; Ülker, B. Precision Genetic Modifications: A New Era in Molecular Biology and Crop Improvement. Planta 2014, 239, 921–939. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An Updated Evolutionary Classification of CRISPR-Cas Systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–822. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 System for Plant Genome Editing and Beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Wyman, C.; Kanaar, R. DNA Double-Strand Break Repair: All’s Well That Ends Well. Annu. Rev. Genet. 2006, 40, 363–383. [Google Scholar] [CrossRef]
- Podevin, N.; Davies, H.V.; Hartung, F.; Nogué, F.; Casacuberta, J.M. Site-Directed Nucleases: A Paradigm Shift in Predictable, Knowledge-Based Plant Breeding. Trends Biotechnol. 2013, 31, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Gao, C. Targeted Genome Modification Technologies and Their Applications in Crop Improvements. Plant Cell Rep. 2014, 33, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 System for Plant Genome Editing: Current Approaches and Emerging Developments. Agronomy 2020, 10, 1033. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two Distinct RNase Activities of CRISPR-C2c2 Enable Guide-RNA Processing and RNA Detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA Targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA Virus Interference via CRISPR/Cas13a System in Plants. Genome Biol. 2018, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Kancharla, N.; Javalkote, V.S.; Dasgupta, S.; Brutnell, T.P. CRISPR-Cas12a (Cpf1): A Versatile Tool in the Plant Genome Editing Tool Box for Agricultural Advancement. Front. Plant Sci. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Endo, A.; Masafumi, M.; Kaya, H.; Toki, S. Efficient Targeted Mutagenesis of Rice and Tobacco Genomes Using Cpf1 from Francisella Novicida. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/DCas9 Platforms in Plants: Strategies and Applications beyond Genome Editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.F. A Potent Cas9-Derived Gene Activator for Plant and Mammalian Cells. Nat. Plants 2017, 3, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust Transcriptional Activation in Plants Using Multiplexed CRISPR-Act2.0 and MTALE-Act Systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef]
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for Highly Efficient Multiplexed Gene Activation in Plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef]
- Malzahn, A.; Zhang, Y.; Qi, Y. CRISPR-Act2.0: An Improved Multiplexed System for Plant Transcriptional Activation. In Plant Genome Editing with CRISPR Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 83–93. [Google Scholar]
- Piatek, A.A.; Lenaghan, S.C.; Neal Stewart, C. Advanced Editing of the Nuclear and Plastid Genomes in Plants. Plant Sci. 2018, 273, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Vilar, M.; Bernabé-Orts, J.M.; Fernandez-del-Carmen, A.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. A Modular Toolbox for GRNA-Cas9 Genome Engineering in Plants Based on the GoldenBraid Standard. Plant Methods BioMed Cent. 2016, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Neumann, M.; Duro, D.I.; Schmid, M. CRISPR-Based Tools for Targeted Transcriptional and Epigenetic Regulation in Plants. PLoS ONE 2019, 14, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.; de de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved Drought Stress Tolerance in Arabidopsis by CRISPR/DCas9 Fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in Agriculture and Plant Biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Sretenovic, S.; Liu, G.; Zhong, Z.; Wang, J.; Huang, L.; Tang, X.; Guo, Y.; Liu, L.; Wu, Y.; et al. Improved Plant Cytosine Base Editors with High Editing Activity, Purity, and Specificity. Plant Biotechnol. J. 2021, 19, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise Plant Genome Editing Using Base Editors and Prime Editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime Genome Editing in Rice and Wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid Generation of a Transgene-Free Powdery Mildew Resistant Tomato by Genome Deletion. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef]
- Dong, O.X.; Yu, S.; Jain, R.; Zhang, N.; Duong, P.Q.; Butler, C.; Li, Y.; Lipzen, A.; Martin, J.A.; Barry, K.W.; et al. Marker-Free Carotenoid-Enriched Rice Generated through Targeted Gene Insertion Using CRISPR-Cas9. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. Gene-Edited CRISPR Mushroom Escapes US Regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [PubMed]
- Globus, R.; Qimron, U. A Technological and Regulatory Outlook on CRISPR Crop Editing. J. Cell. Biochem. 2018, 119, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Teng, F.; Li, T.; Zhou, Q. Simultaneous Generation and Germline Transmission of Multiple Gene Mutations in Rat Using CRISPR-Cas Systems. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient Genome Editing in Plants Using a CRISPR/Cas System. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef]
- Holton, N.; Nekrasov, V.; Ronald, P.C.; Zipfel, C. The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots. PLoS Pathog. 2015, 11, 1–22. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-Guided Genome Editing for Target Gene Mutations in Wheat. G3 Genes Genomes Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome Editing in Rice and Wheat Using the CRISPR/Cas System. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea Mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lipppman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Wang, W.; Xiong, X.; Meng, F.; Cui, X. Efficient Targeted Mutagenesis in Potato by the CRISPR/Cas9 System. Plant Cell Rep. 2015, 34, 1473–1476. [Google Scholar] [CrossRef]
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS ONE 2015, 10, 1–12. [Google Scholar] [CrossRef]
- Michno, J.M.; Wang, X.; Liu, J.; Curtin, S.J.; Kono, T.J.; Stupar, R.M. CRISPR/Cas Mutagenesis of Soybean and Medicago Truncatula Using a New Web-Tool and a Modified Cas9 Enzyme. GM Crops Food 2015, 6, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, T.B.; LaFayette, P.R.; Schmitz, R.J.; Parrott, W.A. Targeted Genome Modifications in Soybean with CRISPR/Cas9. BMC Biotechnol. 2015, 15, 1–10. [Google Scholar] [CrossRef]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted Mutagenesis in Soybean Using the CRISPR-Cas9 System. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-Mediated Genome Editing in Soybean Hairy Roots. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dey, A. CRISPR/Cas Genome Editing to Optimize Pharmacologically Active Plant Natural Products. Pharmacol. Res. 2021, 164, 105359. [Google Scholar] [CrossRef]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome Edited Crops Touch the Market: A View on the Global Development and Regulatory Environment. Front. Plant Sci. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Liu, C.; Cao, Y.; Hua, Y.; Du, G.; Liu, Q.; Wei, X.; Sun, T.; Lin, J.; Wu, M.; Cheng, Z.; et al. Concurrent Disruption of Genetic Interference and Increase of Genetic Recombination Frequency in Hybrid Rice Using CRISPR/Cas9. Front. Plant Sci. 2021, 12, 757152. [Google Scholar] [CrossRef] [PubMed]
- Mieulet, D.; Aubert, G.; Bres, C.; Klein, A.; Droc, G.; Vieille, E.; Rond-Coissieux, C.; Sanchez, M.; Dalmais, M.; Mauxion, J.P.; et al. Unleashing Meiotic Crossovers in Crops. Nat. Plants 2018, 4, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.C.; Alabdullah, A.K.; Moore, G. A Separation-of-Function ZIP4 Wheat Mutant Allows Crossover between Related Chromosomes and Is Meiotically Stable. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zeng, X.; Li, J.; Zhao, D. Construction and Analysis of Tify1a and Tify1b Mutants in Rice (Oryza Sativa) Based on CRISPR/Cas9 Technology. J. Agric. Biotechnol. 2017, 25, 1003–1012. [Google Scholar]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE 2016, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel Alleles of Rice EIF4G Generated by CRISPR/Cas9-Targeted Mutagenesis Confer Resistance to Rice Tungro Spherical Virus. Plant Biotechnol. J. 2018, 16, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Abe, F.; Haque, E.; Hisano, H.; Tanaka, T.; Kamiya, Y.; Mikami, M.; Kawaura, K.; Endo, M.; Onishi, K.; Hayashi, T.; et al. Genome-Edited Triple-Recessive Mutation Alters Seed Dormancy in Wheat. Cell Rep. 2019, 28, 1362–1369.e4. [Google Scholar] [CrossRef]
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D. Multiplex CRISPR/Cas9-Mediated Metabolic Engineering Increases Soya Bean Isoflavone Content and Resistance to Soya Bean Mosaic Virus. Plant Biotechnol. J. 2020, 18, 1384–1395. [Google Scholar] [CrossRef]
- Dong, H.; Huang, Y.; Wang, K. The Development of Herbicide Resistance Crop Plants Using Crispr/Cas9-Mediated Gene Editing. Genes 2021, 12, 912. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient Disruption and Replacement of an Effector Gene in the Oomycete Phytophthora Sojae Using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.E.; Kim, J.G.; Fischer, C.R.; Mehta, N.; Dufour-Schroif, C.; Wemmer, K.; Mudgett, M.B.; Sattely, E. A Pathogen-Responsive Gene Cluster for Highly Modified Fatty Acids in Tomato. Cell 2020, 180, 176–187.e19. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, L.; Zhao, R.; Sheng, J.; Zhang, S.; Li, R.; Shen, L. Knockout of SlMAPK3 Enhances Tolerance to Heat Stress Involving ROS Homeostasis in Tomato Plants. BMC Plant Biol. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-Related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid Improvement of Grain Weight via Highly Efficient CRISPR/Cas9-Mediated Multiplex Genome Editing in Rice. J. Genet. Genom. 2016, 43, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a Subfamily of Abscisic Acid Recepto Genes Promote Rice Growth and Productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing Grain-Yield-Related Traits by CRISPR–Cas9 Promoter Editing of Maize CLE Genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. Cris. J. 2018, 1, 65–74. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and Transgene-Free Genome Editing in Wheat through Transient Expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Wang, W.; Pan, Q.; Tian, B.; He, F.; Chen, Y.; Bai, G.; Akhunova, A.; Trick, H.N.; Akhunov, E. Gene Editing of the Wheat Homologs of TONNEAU1-Recruiting Motif Encoding Gene Affects Grain Shape and Weight in Wheat. Plant J. 2019, 100, 251–264. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, L.; Chen, L.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Yuan, S.; et al. Mutagenesis of GmFT2a and GmFT5a Mediated by CRISPR/Cas9 Contributes for Expanding the Regional Adaptability of Soybean. Plant Biotechnol. J. 2020, 18, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef] [PubMed]
- Akama, K.; Akter, N.; Endo, H.; Kanesaki, M.; Endo, M.; Toki, S. An In Vivo Targeted Deletion of the Calmodulin-Binding Domain from Rice Glutamate Decarboxylase 3 (OsGAD3) Increases γ-Aminobutyric Acid Content in Grains. Rice 2020, 13, 20. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of High-Amylose Rice through CRISPR/Cas9-Mediated Targeted Mutagenesis of Starch Branching Enzymes. Front. Plant Sci. 2021, 8, 1–15. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.; Zhang, C.; Chu, R.; Gu, Z.; Tan, H.; Zhao, D.; Fan, X.; Liu, Q. Creating Novel Wx Alleles with Fine-Tuned Amylose Levels and Improved Grain Quality in Rice by Promoter Editing Using CRISPR/Cas9 System. Plant Biotechnol. J. 2020, 18, 2164–2166. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R.; Lenderts, B.; Yang, M.; Schroder, M.; Farrell, J.; Snopek, K.; Peterson, D.; Feigenbutz, L.; et al. Superior Field Performance of Waxy Corn Engineered Using CRISPR–Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef]
- Ibrahim, S.; Saleem, B.; Rehman, N.; Zafar, S.A.; Naeem, M.K.; Khan, M.R. CRISPR/Cas9 Mediated Disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) Reduces Phytic Acid and Improves Iron and Zinc Accumulation in Wheat Grains. J. Adv. Res. 2021; in press. [Google Scholar] [CrossRef]
- Li, J.; Jiao, G.; Sun, Y.; Chen, J.; Zhong, Y.; Yan, L.; Jiang, D.; Ma, Y.; Xia, L. Modification of Starch Composition, Structure and Properties through Editing of TaSBEIIa in Both Winter and Spring Wheat Varieties by CRISPR/Cas9. Plant Biotechnol. J. 2021, 19, 937–951. [Google Scholar] [CrossRef]
- Sánchez-León, S.; Gil-Humanes, J.; Ozuna, C.V.; Giménez, M.J.; Sousa, C.; Voytas, D.F.; Barro, F. Low-Gluten, Nontransgenic Wheat Engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Camerlengo, F.; Frittelli, A.; Sparks, C.; Doherty, A.; Martignago, D.; Larré, C.; Lupi, R.; Sestili, F.; Masci, S. CRISPR-Cas9 Multiplex Editing of the α-Amylase/Trypsin Inhibitor Genes to Reduce Allergen Proteins in Durum Wheat. Front. Sustain. Food Syst. 2020, 4, 104. [Google Scholar] [CrossRef]
- Raffan, S.; Sparks, C.; Huttly, A.; Hyde, L.; Martignago, D.; Mead, A.; Hanley, S.J.; Wilkinson, P.A.; Barker, G.; Edwards, K.J.; et al. Wheat with Greatly Reduced Accumulation of Free Asparagine in the Grain, Produced by CRISPR/Cas9 Editing of Asparagine Synthetase Gene TaASN2. Plant Biotechnol. J. 2021, 19, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A.; Corbin, K.R.; Ahn-Jarvis, J.; Harris, S.; Hawkins, E.; Smedley, M.A.; Harwood, W.; Warren, F.J.; Patron, N.J.; Smith, A.M. Cas9-Mediated Mutagenesis of Potato Starch-Branching Enzymes Generates a Range of Tuber Starch Phenotypes. Plant Biotechnol. J. 2019, 17, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Y.; Zhang, Y.; Yang, J.; Fan, W.; Zhang, H.; Zhao, S.; Yuan, L.; Zhang, P. CRISPR/Cas9-Based Mutagenesis of Starch Biosynthetic Genes in Sweet Potato (Ipomoea Batatas) for the Improvement of Starch Quality. Int. J. Mol. Sci. 2019, 20, 4702. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Nonaka, S.; Arai, C.; Takayama, M.; Matsukura, C.; Ezura, H. Efficient Increase of Γ-Aminobutyric Acid (GABA) Content in Tomato Fruits by Targeted Mutagenesis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Li, R.; Li, R.; Li, X.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. Multiplexed CRISPR/Cas9-Mediated Metabolic Engineering of γ-Aminobutyric Acid Levels in Solanum Lycopersicum. Plant Biotechnol. J. 2018, 16, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Blennow, A.; Kofoed-Enevoldsen, O.; Jiang, D.; Hebelstrup, K.H. Protein Targeting to Starch 1 Is Essential for Starchy Endosperm Development in Barley. J. Exp. Bot. 2019, 70, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhong, X.; Li, Q.; Lan, J.; Tang, H.; Qi, P.; Ma, J.; Wang, J.; Chen, G.; Pu, Z.; et al. Mutation of the D-Hordein Gene by RNA-Guided Cas9 Targeted Editing Reducing the Grain Size and Changing Grain Compositions in Barley. Food Chem. 2020, 311, 125892. [Google Scholar] [CrossRef] [PubMed]
- Sashidhar, N.; Harloff, H.J.; Potgieter, L.; Jung, C. Gene Editing of Three BnITPK Genes in Tetraploid Oilseed Rape Leads to Significant Reduction of Phytic Acid in Seeds. Plant Biotechnol. J. 2020, 18, 2241–2250. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.S.; Samuelsson, M.; Hofvander, P. Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum tuberosum) by Transient CRISPR-Cas9 Expression in Protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Hayut, S.F.; Bessudo, C.M.; Levy, A.A. Targeted Recombination between Homologous Chromosomes for Precise Breeding in Tomato. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Shlush, I.; Ben Samach, A.; Melamed-Bessudo, C.; Ben-Tov, D.; Dahan-Meir, T.; Filler-Hayut, S.; Levy, A.A. Crispr/Cas9 Induced Somatic Recombination at the Crtiso Locus in Tomato. Forests 2021, 12, 59. [Google Scholar] [CrossRef]
- Lee, J.H.; Won, H.J.; Hoang Nguyen Tran, P.; Lee, S.M.; Kim, H.Y.; Jung, J.H. Improving Lignocellulosic Biofuel Production by CRISPR/Cas9-mediated Lignin Modification in Barley. GCB Bioenergy 2021, 13, 742–752. [Google Scholar] [CrossRef]
- Beying, N.; Schmidt, C.; Pacher, M.; Houben, A.; Puchta, H. CRISPR–Cas9-Mediated Induction of Heritable Chromosomal Translocations in Arabidopsis. Nat. Plants 2020, 6, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Park, J.J.; Yoo, C.G.; Flanagan, A.; Pu, Y.; Debnath, S.; Ge, Y.; Ragauskas, A.J.; Wang, Z.Y. Defined Tetra-Allelic Gene Disruption of the 4-Coumarate:Coenzyme A Ligase 1 (Pv4CL1) Gene by CRISPR/Cas9 in Switchgrass Results in Lignin Reduction and Improved Sugar Release Mike Himmel. Biotechnol. Biofuels BioMed Cent. 2017, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, A.; Ogawa, T.; Koizuka, C.; Kaneko, K.; Inaba, M.; Imamura, J.; Koizuka, N. CRISPR/Cas9-Mediated Genome Editing of the Fatty Acid Desaturase 2 Gene in Brassica Napus. Plant Physiol. Biochem. 2018, 131, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cui, G.; Shen, G.; Zhan, Z.; Huang, L.; Chen, J.; Qi, X. Targeted Mutagenesis in the Medicinal Plant Salvia Miltiorrhiza. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Liu, X.; Fan, C.; Li, T.; Qin, H.; Li, X.; Chen, K.; Zheng, Y.; Chen, F.; Xu, Y. Enhancement of Tobacco (Nicotiana Tabacum L.) Seed Lipid Content for Biodiesel Production by CRISPR-Cas9-Mediated Knockout of NtAn1. Front. Plant Sci. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Zhang, J.; Botella, J.R.; Ma, C.; Kong, F.; Liu, B.; Zhu, J.K. Perspectives on the Application of Genome-Editing Technologies in Crop Breeding. Mol. Plant 2019, 12, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and Application of Plant Mutagenesis in Crop Improvement: A Review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef]
- Chaudhary, J.; Deshmukh, R.; Sonah, H. Mutagenesis Approaches and Their Role in Crop Improvement. Plants 2019, 8, 467. [Google Scholar] [CrossRef]
- Raina, A.; Laskar, R.; Khursheed, S.; Amin, R.; Tantray, Y.; Parveen, K.; Khan, S. Role of Mutation Breeding in Crop Improvement- Past, Present and Future. Asian Res. J. Agric. 2016, 2, 1–13. [Google Scholar] [CrossRef]
- Sharma, S.; Kaur, R.; Singh, A. Recent Advances in CRISPR/Cas Mediated Genome Editing for Crop Improvement. Plant Biotechnol. Rep. 2017, 11, 193–207. [Google Scholar] [CrossRef]
- Aglawe, S.B.; Barbadikar, K.M.; Mangrauthia, S.K.; Madhav, M.S. New Breeding Technique “Genome Editing” for Crop Improvement: Applications, Potentials and Challenges. Biotech 2018, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Filler-hayut, S.; Kniazev, K.; Melamed-bessudo, C.; Levy, A.A. Targeted Inter-Homologs Recombination in Arabidopsis Euchromatin and Heterochromatin. Int. J. Mol. Sci. 2021, 22, 12096. [Google Scholar] [CrossRef] [PubMed]
- Taagen, E.; Bogdanove, A.J.; Sorrells, M.E. Counting on Crossovers: Controlled Recombination for Plant Breeding. Trends Plant Sci. 2020, 25, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Blary, A.; Jenczewski, E. Manipulation of Crossover Frequency and Distribution for Plant Breeding. Theor. Appl. Genet. 2019, 132, 575–592. [Google Scholar] [CrossRef]
- Schaart, J.G.; van de Wiel, C.C.M.; Lotz, L.A.P.; Smulders, M.J.M. Opportunities for Products of New Plant Breeding Techniques. Trends Plant Sci. 2016, 21, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Gill, R.A.; Jung, K.H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and Molecular Techniques from Simple Breeding to Speed Breeding in Crop Plants: Recent Advances and Future Outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef] [PubMed]
- Qaim, M. Role of New Plant Breeding Technologies for Food Security and Sustainable Agricultural Development. Appl. Econ. Perspect. Policy 2020, 42, 129–150. [Google Scholar] [CrossRef]
- Hartung, F.; Schiemann, J. Precise Plant Breeding Using New Genome Editing Techniques: Opportunities, Safety and Regulation in the EU. Plant J. 2014, 78, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Abdel Latef, A.A.H.; Rasool, S.; Akram, N.A.; Ashraf, M.; Gucel, S. Role of Proteomics in Crop Stress Tolerance. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hussain, B. Modernization in Plant Breeding Approaches for Improving Biotic Stress Resistance in Crop Plants. Turkish. J. Agric. For. 2015, 39, 515–530. [Google Scholar] [CrossRef]
- Bigini, V.; Camerlengo, F.; Botticella, E.; Sestili, F.; Savatin, D.V. Biotechnological Resources to Increase Disease-Resistance by Improving Plant Immunity: A Sustainable Approach to Save Cereal Crop Production. Plants 2021, 10, 1146. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Shan-e-Ali Zaidi, S.; Vanderschuren, H.; Qaim, M.; Mahfouz, M.M.; Kohli, A.; Mansoor, S.; Tester, M. New Plant Breeding Technologies for Food Security. Science 2019, 363, 1390–1391. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Meng, X.; Liu, Q.; Li, J.; Wang, K. Increasing the Efficiency of CRISPR-Cas9-VQR Precise Genome Editing in Rice. Plant Biotechnol. J. 2018, 16, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome Editing in Rice: Recent Advances, Challenges, and Future Implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013, 41, 1–12. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted Mutagenesis in Rice Using CRISPR-Cas System. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-Guided Genome Editing in Plants Using a CRISPR-Cas System. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Farhat, S.; Jain, N.; Singh, N.; Sreevathsa, R.; Dash, P.K.; Rai, R.; Yadav, S.; Kumar, P.; Sarkar, A.K.; Jain, A.; et al. CRISPR-Cas9 Directed Genome Engineering for Enhancing Salt Stress Tolerance in Rice. Semin. Cell Dev. Biol. 2019, 96, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Kihoro, J.; Bosco, N.J.; Murage, H.; Ateka, E.; Makihara, D. Investigating the Impact of Rice Blast Disease on the Livelihood of the Local Farmers in Greater Mwea Region of Kenya. Springerplus 2013, 2, 1–13. [Google Scholar] [CrossRef]
- Sánchez, F.J.; Arciniegas, J.P.; Brand, A.; Vacca, O.; Tohme, J.; Becerra, L.A. E Dición de Genomas En Manihot Esculenta Crantz Para La Producción de Plantas Waxy y Para La Resistencia a La Bacteriosis Vascular Producida Por Xanthomonas Axonopodis P. In Proceedings of the X Encuentro Latinoamericano y Del Caribe de Biotecnología Agropecuaria y XI Simposio REDBIO Argentina, Montevideo, Uruguay, 12–15 November 2019. [Google Scholar] [CrossRef]
- Tokatlidis, I.S. Adapting Maize Crop to Climate Change. Agron. Sustain. Dev. 2013, 33, 63–79. [Google Scholar] [CrossRef]
- Sangoi, L.; Gracietti, M.A.; Rampazzo, C.; Bianchetti, P. Response of Brazilian Maize Hybrids from Different Eras to Changes in Plant Density. F Crop. Res. 2002, 79, 39–51. [Google Scholar] [CrossRef]
- Gatica-Arias, A. The Regulatory Current Status of Plant Breeding Technologies in Some Latin American and the Caribbean Countries. Plant Cell. Tissue Organ Cult. 2020, 141, 229–242. [Google Scholar] [CrossRef]
- Kumar, R.; Kaur, A.; Pandey, A.; Mamrutha, H.M.; Singh, G.P. CRISPR-Based Genome Editing in Wheat: A Comprehensive Review and Future Prospects. Mol. Biol. Rep. 2019, 46, 3557–3569. [Google Scholar] [CrossRef]
- Khojely, D.M.; Ibrahim, S.E.; Sapey, E.; Han, T. History, Current Status, and Prospects of Soybean Production and Research in Sub-Saharan Africa. Crop J. 2018, 6, 226–235. [Google Scholar] [CrossRef]
- Das Dangol, S.; Barakate, A.; Stephens, J.; Çalıskan, M.E.; Bakhsh, A. Genome Editing of Potato Using CRISPR Technologies: Current Development and Future Prospective. Plant Cell. Tissue Organ Cult. 2019, 139, 403–416. [Google Scholar] [CrossRef]
- Dobrovidova, O. Russia Joins in Global Gene-Editing Bonanza. Nature 2019, 569, 319–320. [Google Scholar] [CrossRef]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in Application of Genome Editing in Tomato and Recent Development of Genome Editing Technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A Single Transcription Factor Promotes Both Yield and Immunity in Rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Sommer, A. Vitamin A Deficiency and Clinical Disease: An Historical Overview. J. Nutr. 2008, 138, 1835–1839. [Google Scholar] [CrossRef]
- Sparvoli, F.; Cominelli, E. Seed Biofortification and Phytic Acid Reduction: A Conflict of Interest for the Plant? Plants 2015, 4, 728–755. [Google Scholar] [CrossRef]
- Mottram, D.S.; Wedzicha, B.L.; Dodson, A.T. Food Chemistry: Acrylamide Is Formed in the Maillard Reaction. Nature 2002, 419, 448–449. [Google Scholar] [CrossRef]
- Regina, A.; Bird, A.; Topping, D.; Bowden, S.; Freeman, J.; Barsby, T.; Kosar-Hashemi, B.; Li, Z.; Rahman, S.; Morell, M. High-Amylose Wheat Generated by RNA Interference Improves Indices of Large-Bowel Health in Rats. Proc. Natl. Acad. Sci. USA 2006, 103, 3546–3551. [Google Scholar] [CrossRef]
- Tatham, A.S.; Shewry, P.R. Allergens to Wheat and Related Cereals. Clin. Exp. Allergy 2008, 38, 1712–1726. [Google Scholar] [CrossRef]
- Mansueto, P.; Soresi, M.; Iacobucci, R.; La Blasca, F.; Romano, G.; D’Alcamo, A.; Carroccio, A. Non-Celiac Wheat Sensitivity: A Search for the Pathogenesis of a Self-Reported Condition. Ital. J. Med. 2019, 13, 15–23. [Google Scholar] [CrossRef]
- Geisslitz, S.; Shewry, P.; Brouns, F.; America, A.H.P.; Caio, G.P.I.; Daly, M.; D’Amico, S.; De Giorgio, R.; Gilissen, L.; Grausgruber, H.; et al. Wheat ATIs: Characteristics and Role in Human Disease. Front. Nutr. 2021, 8, 1–16. [Google Scholar] [CrossRef]
- Schmidt, C.; Schindele, P.; Puchta, H. From Gene Editing to Genome Engineering: Restructuring Plant Chromosomes via CRISPR/Cas. aBiotech 2020, 1, 21–31. [Google Scholar] [CrossRef]
- Schindele, A.; Dorn, A.; Puchta, H. CRISPR/Cas Brings Plant Biology and Breeding into the Fast Lane. Curr. Opin. Biotechnol. 2020, 61, 7–14. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large Chromosomal Deletions and Heritable Small Genetic Changes Induced by CRISPR/Cas9 in Rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Lenderts, B.; Feigenbutz, L.; Barone, P.; Llaca, V.; Fengler, K.; Svitashev, S. CRISPR–Cas9-mediated 75.5-Mb inversion in maize. Nat. Plants 2020, 6, 1427–1431. [Google Scholar] [CrossRef]
- Huang, D.; Kosentka, P.Z.; Liu, W. Synthetic Biology Approaches in Regulation of Targeted Gene Expression. Curr. Opin. Plant Biol. 2021, 63, 102036. [Google Scholar] [CrossRef]
- Karlson, C.K.S.; Mohd-noor, S.N.; Nolte, N.; Tan, B.C. Crispr/Dcas9-based Systems: Mechanisms and Applications in Plant Sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Si, X.; Zhang, H.; Wang, Y.; Chen, K.; Gao, C. Manipulating Gene Translation in Plants by CRISPR–Cas9-Mediated Genome Editing of Upstream Open Reading Frames. Nat. Protoc. 2020, 15, 338–363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome Editing of Upstream Open Reading Frames Enables Translational Control in Plants. Nat. Biotechnol. 2018, 36, 894–900. [Google Scholar] [CrossRef]
- Schachtsiek, J.; Stehle, F. Nicotine-Free, Nontransgenic Tobacco (Nicotiana Tabacum l.) Edited by CRISPR-Cas9. Plant Biotechnol. J. 2019, 17, 2228–2230. [Google Scholar] [CrossRef]
- Morris, S.H.; Spillane, C. GM Directive Deficiencies in the European Union. The Current Framework for Regulating GM Crops in the EU Weakens the Precautionary Principle as a Policy Tool. EMBO Rep. 2008, 9, 500–504. [Google Scholar] [CrossRef]
- Podevin, N.; Devos, Y.; Davies, H.V.; Nielsen, K.M. Transgenic or Not? No Simple Answer! New Biotechnology-Based Plant Breeding Techniques and the Regulatory Landscape. EMBO Rep. 2012, 13, 1057–1061. [Google Scholar] [CrossRef]
- Heap, B. Europe Should Rethink Its Stance on GM Crops. Nature 2013, 498, 409. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camerlengo, F.; Frittelli, A.; Pagliarello, R. CRISPR towards a Sustainable Agriculture. Encyclopedia 2022, 2, 538-558. https://0-doi-org.brum.beds.ac.uk/10.3390/encyclopedia2010036
Camerlengo F, Frittelli A, Pagliarello R. CRISPR towards a Sustainable Agriculture. Encyclopedia. 2022; 2(1):538-558. https://0-doi-org.brum.beds.ac.uk/10.3390/encyclopedia2010036
Chicago/Turabian StyleCamerlengo, Francesco, Arianna Frittelli, and Riccardo Pagliarello. 2022. "CRISPR towards a Sustainable Agriculture" Encyclopedia 2, no. 1: 538-558. https://0-doi-org.brum.beds.ac.uk/10.3390/encyclopedia2010036