A COVID-19 Overview and Potential Applications of Cell Therapy
Abstract
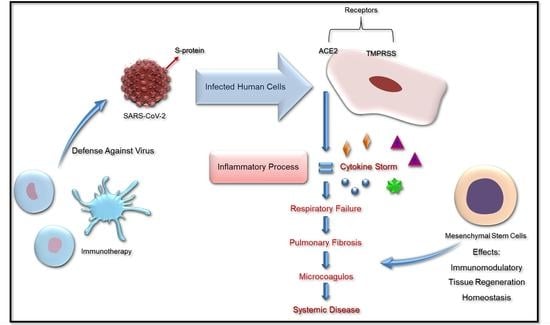
:1. Introduction
2. SARS-CoV-2 Structure
3. Spike Glycoprotein and ACE2 Receptor Interaction
4. Hematologic Disease
5. Stem Cell Treatment
6. Immunotherapy Based in NK Cells
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nat. Cell Biol. 2016, 531, 118–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Novel Coronavirus (2019-nCoV) Situation Report; WHO Bull. 2020. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200123-sitrep-3-2019-ncov.pdf (accessed on 11 June 2021).
- Velavan, T.P.; Meyer, C.G. The COVID-19 epidemic. Trop. Med. Int. Health 2020, 25, 278–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Tsang, K.W.; Ho, P.L.; Ooi, G.C.; Yee, W.K.; Wang, T.; Chan-Yeung, M.; Lam, W.K.; Seto, W.H.; Yam, L.Y.; Cheung, T.M.; et al. A Cluster of Cases of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1977–1985. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020, 20, 425–434. [Google Scholar] [CrossRef]
- Galván Casas, C.; Català, A.; Carretero Hernández, G.; Rodríguez-Jiménez, P.; Fernández-Nieto, D.; Rodríguez-Villa Lario, A.; Navarro Fernández, I.; Ruiz-Villaverde, R.; Falkenhain-López, D.; Llamas Velasco, M.; et al. Classification of the Cutaneous Manifestations of COVID-19: A Rapid Prospective Nationwide Consensus Study in Spain with 375 Cases. Br. J. Dermatol. 2020, 183, 71–77. [Google Scholar] [CrossRef]
- Lake, M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin. Med. 2020, 20, 124–127. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Mortola, E.; Roy, P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004, 576, 174–178. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Álvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene Is Attenuated In Vitro and In Vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar] [CrossRef] [Green Version]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Kuo, L.; Hurst, K.R.; Masters, P.S. Exceptional Flexibility in the Sequence Requirements for Coronavirus Small Envelope Protein Function. J. Virol. 2007, 81, 2249–2262. [Google Scholar] [CrossRef] [Green Version]
- Neuman, B.W.; Kiss, G.; Kunding, A.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017, 8, 15092. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessa, T.; Kim, H.; Bihlmaier, K.; Lundin, C.; Boekel, C.; Andersson, H.; Nilsson, I.; White, S.; von Heijne, G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nat. Cell Biol. 2005, 433, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, J.; Zhou, Y.; Hua, J.; Zhang, L.; Bian, J.; Liu, B.; Zhao, Z.; Jin, S. The scRNA-seq expression profiling of the receptor ACE2 and the cellular protease TMPRSS2 reveals human organs susceptible to COVID-19 infection. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A Transmembrane Serine Protease Is Linked to the Severe Acute Respiratory Syndrome Coronavirus Receptor and Activates Virus Entry. J. Virol. 2010, 85, 873–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273, Addendum in 2020, 588, E6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, W.; Li, T. COVID-19: Towards understanding of pathogenesis. Cell Res. 2020, 30, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [Green Version]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Elalamy, I.; Kastritis, E.; Sergentanis, T.N.; Politou, M.; Psaltopoulou, T.; Gerotziafas, G.; Dimopoulos, M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020, 95, 834–847. [Google Scholar] [CrossRef] [Green Version]
- Gattinoni, L.; Coppola, S.; Cressoni, M.; Busana, M.; Rossi, S.; Chiumello, D. COVID-19 Does Not Lead to a “Typical” Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2020, 201, 1299–1300. [Google Scholar] [CrossRef] [Green Version]
- Subbarao, K.; Mahanty, S. Respiratory Virus Infections: Understanding COVID-19. Immunity 2020, 52, 905–909. [Google Scholar] [CrossRef]
- Poor, H.D.; Ventetuolo, C.E.; Tolbert, T.; Chun, G.; Serrao, G.; Zeidman, A.; Dangayach, N.S.; Olin, J.; Kohli-Seth, R.; Powell, C.A. COVID-19 critical illness pathophysiology driven by diffuse pulmonary thrombi and pulmonary endothelial dysfunction responsive to thrombolysis. Clin. Transl. Med. 2020, 10, 2–6. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.; Bai, H.; Chen, X.; Gong, J.; Li, D.; Sun, Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020, 18, 1094–1099. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.-Q.; Wang, Q.; Miao, H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct. Target. Ther. 2020, 5, 1–3. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Thachil, J.; Tang, N.; Gando, S.; Falanga, A.; Cattaneo, M.; Levi, M.; Clark, C.; Iba, T. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J. Thromb. Haemost. 2020, 18, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Debuc, B.; Smadja, D.M. Is COVID-19 a New Hematologic Disease? Stem Cell Rev. Rep. 2021, 17, 4–8. [Google Scholar] [CrossRef]
- Shi, Y.; Su, J.; Roberts, A.I.; Shou, P.; Rabson, A.B.; Ren, G. How mesenchymal stem cells interact with tissue immune responses. Trends Immunol. 2012, 33, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.-J.; Kim, N.; Cho, S.-G. The potential use of mesenchymal stem cells in hematopoietic stem cell transplantation. Exp. Mol. Med. 2013, 45, e2. [Google Scholar] [CrossRef] [Green Version]
- Zachar, L.; Bačenková, D.; Rosocha, J. Activation, homing, and role of the mesenchymal stem cells in the inflammatory environment. J. Inflamm. Res. 2016, 9, 231–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsonraj, R.M.; Raghunath, M.; Nurcombe, V.; Hui, J.H.; Van Wijnen, A.J.; Cool, S.M. Concise Review: Multifaceted Characterization of Human Mesenchymal Stem Cells for Use in Regenerative Medicine. Stem Cells Transl. Med. 2017, 6, 2173–2185. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.H.; Kim, H.J.; Jeong, H.J.; Lee, H.J.; Oh, J.Y. Mesenchymal Stem and Stromal Cells Harness Macrophage-Derived Amphiregulin to Maintain Tissue Homeostasis. Cell Rep. 2020, 30, 3806–3820.e6. [Google Scholar] [CrossRef] [Green Version]
- Alfaifi, M.; Eom, Y.W.; Newsome, P.N.; Baik, S.K. Mesenchymal stromal cell therapy for liver diseases. J. Hepatol. 2018, 68, 1272–1285. [Google Scholar] [CrossRef] [Green Version]
- Falkenburg, F.; Ruggiero, E.; Bonini, M.C.; Porter, D.; Miller, J.; Malard, F.; Mohty, M.; Kröger, N.; Kolb, H.J. Prevention and treatment of relapse after stem cell transplantation by cellular therapies. Bone Marrow Transpl. 2018, 54, 26–34. [Google Scholar] [CrossRef]
- Jiang, W.; Xu, J. Immune modulation by mesenchymal stem cells. Cell Prolif. 2019, 53, e12712. [Google Scholar] [CrossRef]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2- Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020, 11, 216–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.W.; Fang, X.; Krasnodembskaya, A.; Howard, J.P.; Matthay, M.A. Concise review: Mesenchymal stem cells for acute lung injury: role of paracrine soluble factors. Stem Cells 2011, 29, 913–919. [Google Scholar] [CrossRef] [Green Version]
- Golchin, A.; Seyedjafari, E.; Ardeshirylajimi, A. Mesenchymal Stem Cell Therapy for COVID-19: Present or Future. Stem Cell Rev. Rep. 2020, 16, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Cruz, F.F.; Rocco, P.R.M. The potential of mesenchymal stem cell therapy for chronic lung disease. Expert Rev. Respir. Med. 2020, 14, 31–39. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Bonanni, V.; Sciume, G.; Santoni, A.; Bernardini, G. Bone Marrow NK Cells: Origin, Distinctive Features, and Requirements for Tissue Localization. Front. Immunol. 2019, 10, 1569. [Google Scholar] [CrossRef] [Green Version]
- Zucchini, N.; Crozat, K.; Baranek, T.; Robbins, S.H.; Altfeld, M.; Dalod, M. Natural killer cells in immunodefense against infective agents. Expert Rev. Anti-Infective Ther. 2008, 6, 867–885. [Google Scholar] [CrossRef]
- Malhotra, A.; Shanker, A. NK cells: Immune cross-talk and therapeutic implications. Immunotherapy 2011, 3, 1143–1166. [Google Scholar] [CrossRef] [Green Version]
- Della Chiesa, M.; De Maria, A.; Muccio, L.; Bozzano, F.; Sivori, S.; Moretta, L. Human NK Cells and Herpesviruses: Mechanisms of Recognition, Response and Adaptation. Front. Microbiol. 2019, 10, 2297. [Google Scholar] [CrossRef]
- Dahlberg, C.I.M.; Sarhan, D.; Chrobok, M.; Duru, A.; Alici, E. Natural Killer Cell-Based Therapies Targeting Cancer: Possible Strategies to Gain and Sustain Anti-Tumor Activity. Front. Immunol. 2015, 6, 605. [Google Scholar] [CrossRef] [Green Version]
- Farag, S.S.; Fehniger, T.; Ruggeri, L.; Velardi, A.; Caligiuri, M.A. Natural killer cell receptors: New biology and insights into the graft-versus-leukemia effect. Blood 2002, 100, 1935–1947. [Google Scholar] [CrossRef]
- Peppa, D. Natural Killer Cells in Human Immunodeficiency Virus-1 Infection: Spotlight on the Impact of Human Cytomegalovirus. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Mandal, A.; Viswanathan, C. Natural killer cells: In health and disease. Hematol. Stem Cell Ther. 2015, 8, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Braun, M.; Björkström, N.; Gupta, S.; Sundström, K.; Ahlm, C.; Klingstrom, J.; Ljunggren, H.-G. NK Cell Activation in Human Hantavirus Infection Explained by Virus-Induced IL-15/IL15Rα Expression. PLoS Pathog. 2014, 10, e1004521. [Google Scholar] [CrossRef] [Green Version]
- Petitdemange, C.; Becquart, P.; Wauquier, N.; Béziat, V.; Debré, P.; Leroy, E.; Vieillard, V. Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity. PLoS Pathog. 2011, 7, e1002268. [Google Scholar] [CrossRef] [Green Version]
- Rehermann, B. Natural Killer Cells in Viral Hepatitis. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 578–588. [Google Scholar] [CrossRef] [Green Version]
- Chijioke, O.; Landtwing, V.; Münz, C. NK Cell Influence on the Outcome of Primary Epstein–Barr Virus Infection. Front. Immunol. 2016, 7, 323. [Google Scholar] [CrossRef] [Green Version]
- Lion, E.; Anguille, S.; Berneman, Z.N.; Smits, E.L.J.M.; Van Tendeloo, V.F.I. Poly(I:C) Enhances the Susceptibility of Leukemic Cells to NK Cell Cytotoxicity and Phagocytosis by DC. PLoS ONE 2011, 6, e20952. [Google Scholar] [CrossRef]
- Smits, E.L.J.M.; Ponsaerts, P.; Van De Velde, A.L.R.; Van Driessche, A.; Cools, N.; Lenjou, M.; Nijs, G.; Van Bockstaele, D.R.; Berneman, Z.N.; Van Tendeloo, V.F.I. Proinflammatory response of human leukemic cells to dsRNA transfection linked to activation of dendritic cells. Leukemia 2007, 21, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Kübler, K.; Pesch, C.T.; Gehrke, N.; Riemann, S.; Daßler, J.; Coch, C.; Landsberg, J.; Wimmenauer, V.; Pölcher, M.; Rudlowski, C.; et al. Immunogenic cell death of human ovarian cancer cells induced by cytosolic poly(I:C) leads to myeloid cell maturation and activates NK cells. Eur. J. Immunol. 2011, 41, 3028–3039. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.; Davis, J.E.; Grigg, A.; Szer, J.; Ritchie, D. Strategies to enhance the graft versus tumour effect after allogeneic haematopoietic stem cell transplantation. Bone Marrow Transpl. 2018, 54, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Yeap, W.H.; Wong, K.L.; Shimasaki, N.; Teo, E.C.Y.; Quek, J.K.S.; Yong, H.X.; Diong, C.P.; Bertoletti, A.; Linn, Y.C.; Wong, S.C. CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes. Sci. Rep. 2016, 6, 34310. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Câmara, D.A.D.; Porcacchia, A.S.; Lizier, N.F.; De-Sá-Júnior, P.L. A COVID-19 Overview and Potential Applications of Cell Therapy. Biologics 2021, 1, 177-188. https://0-doi-org.brum.beds.ac.uk/10.3390/biologics1020011
Câmara DAD, Porcacchia AS, Lizier NF, De-Sá-Júnior PL. A COVID-19 Overview and Potential Applications of Cell Therapy. Biologics. 2021; 1(2):177-188. https://0-doi-org.brum.beds.ac.uk/10.3390/biologics1020011
Chicago/Turabian StyleCâmara, Diana Aparecida Dias, Allan Saj Porcacchia, Nelson Foresto Lizier, and Paulo Luiz De-Sá-Júnior. 2021. "A COVID-19 Overview and Potential Applications of Cell Therapy" Biologics 1, no. 2: 177-188. https://0-doi-org.brum.beds.ac.uk/10.3390/biologics1020011