Marine Animal-Derived Compounds and Autophagy Modulation in Breast Cancer Cells
Abstract
:1. A Brief Insight into Autophagy and Breast Cancer Cells
2. The Marine Animal Species as a Source of Bioactive Molecules
3. Autophagy Modulators from Porifera
4. Autophagy Modulators from Cnidaria
5. Autophagy Modulators from Mollusca
6. Autophagy Modulators from Echinodermata
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chun, Y.; Kim, J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells 2018, 7, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caradonna, F.; Cruciata, I.; Luparello, C. Nutrigenetics, nutrigenomics and phenotypic outcomes of dietary low-dose alcohol consumption in the suppression and induction of cancer development: Evidence from in vitro studies. Crit. Rev. Food Sci. Nutr. 2020, 8, 1–32, epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Sarkar, S.; Davies, J.E.; Futter, M.; Garcia-Arencibia, M.; Green-Thompson, Z.W.; Jimenez-Sanchez, M.; Korolchuk, V.I.; Lichtenberg, M.; Luo, S.; et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010, 90, 1383–1435. [Google Scholar] [CrossRef] [Green Version]
- Al-Bari, M.A.A.; Xu, P. Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann. N. Y. Acad. Sci. 2020, 1467, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 8, 1–382. [Google Scholar] [CrossRef]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef] [Green Version]
- Lazova, R.; Camp, R.L.; Klump, V.; Siddiqui, S.F.; Amaravadi, R.K.; Pawelek, J.M. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. 2012, 18, 370–379. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.C.; Wu, A.G.; Huang, Y.Z.; Shao, G.L.; Ji, S.F.; Wang, R.W.; Yuan, H.J.; Fan, X.L.; Zheng, L.H.; Jiao, Q.L. Autophagic regulation of cell growth by altered expression of Beclin 1 in triple-negative breast cancer. Int. J. Clin. Exp. Med. 2015, 8, 7049–7058. [Google Scholar] [PubMed]
- Librizzi, M.; Longo, A.; Chiarelli, R.; Amin, J.; Spencer, J.; Luparello, C. Cytotoxic effects of Jay Amin hydroxamic acid (JAHA), a ferrocene-based class I histone deacetylase inhibitor, on triple-negative MDA-MB231 breast cancer cells. Chem. Res. Toxicol. 2012, 25, 2608–2616. [Google Scholar] [CrossRef] [Green Version]
- Librizzi, M.; Tobiasch, E.; Luparello, C. The conditioned medium from osteo-differentiating human mesenchymal stem cells affects the viability of triple negative MDA-MB231 breast cancer cells. Cell Biochem. Funct. 2016, 34, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Librizzi, M.; Spencer, J.; Luparello, C. Biological Effect of a Hybrid Anticancer Agent Based on Kinase and Histone Deacetylase Inhibitors on Triple-Negative (MDA-MB231) Breast Cancer Cells. Int. J. Mol. Sci. 2016, 17, 1235. [Google Scholar] [CrossRef] [Green Version]
- Luparello, C.; Asaro, D.M.L.; Cruciata, I.; Hassell-Hart, S.; Sansook, S.; Spencer, J.; Caradonna, F. Cytotoxic Activity of the Histone Deacetylase 3-Selective Inhibitor Pojamide on MDA-MB-231 Triple-Negative Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maycotte, P.; Thorburn, A. Targeting autophagy in breast cancer. World J. Clin. Oncol. 2014, 5, 224–240. [Google Scholar] [CrossRef] [PubMed]
- Folkerts, H.; Hilgendorf, S.; Vellenga, E.; Bremer, E.; Wiersma, V.R. The multifaceted role of autophagy in cancer and the microenvironment. Med. Res. Rev. 2019, 39, 517–560. [Google Scholar] [CrossRef] [PubMed]
- Cocco, S.; Leone, A.; Piezzo, M.; Caputo, R.; Di Lauro, V.; Di Rella, F.; Fusco, G.; Capozzi, M.; Gioia, G.d.; Budillon, A.; et al. Targeting Autophagy in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 7836. [Google Scholar] [CrossRef]
- Malve, H. Exploring the ocean for new drug developments: Marine pharmacology. J. Pharm. Bioallied Sci. 2016, 8, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.J.M.; Roque, A.C.A. Free Marine Natural Products Databases for Biotechnology and Bioengineering. Biotechnol. J. 2019, 14, e1800607. [Google Scholar] [CrossRef] [PubMed]
- Mayekar, T.S.; Salgaonkar, A.A.; Koli, J.M.; Patil, P.; Chaudhari, A.; Murkar, A.; Salvi, S.; Surve, D.; Jadhav, R.; Kazi, T. Marine Biotechnology: Bioactive Natural Products and their Applications. Available online: http://aquafind.com/articles/Marine-Biotechnology.php (accessed on 19 March 2021).
- Saha, M.; Berdalet, E.; Carotenuto, Y.; Fink, P.; Harder, T.; John, U.; Not, F.; Pohnert, G.; Potin, P.; Selander, E.; et al. Using chemical language to shape future marine health. Front. Ecol. Environ. 2019, 17, 530–537. [Google Scholar] [CrossRef]
- Lazzara, V.; Arizza, V.; Luparello, C.; Mauro, M.; Vazzana, M. Bright Spots in the Darkness of Cancer: A Review of Starfishes-Derived Compounds and Their Anti-Tumor Action. Mar. Drugs 2019, 17, 617. [Google Scholar] [CrossRef] [Green Version]
- Mauro, M.; Lazzara, V.; Punginelli, D.; Arizza, V.; Vazzana, M. Antitumoral compounds from vertebrate sister group: A review of Mediterranean ascidians. Dev. Comp. Immunol. 2020, 108, 103669. [Google Scholar] [CrossRef]
- Luparello, C.; Mauro, M.; Lazzara, V.; Vazzana, M. Collective Locomotion of Human Cells, Wound Healing and Their Control by Extracts and Isolated Compounds from Marine Invertebrates. Molecules 2020, 25, 2471. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Mauro, M.; Arizza, V.; Vazzana, M. Histone Deacetylase Inhibitors from Marine Invertebrates. Biology 2020, 9, 429. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Doan, C.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.-L. Reclamation of Fishery Processing Waste: A Mini-Review. Molecules 2019, 24, 2234. [Google Scholar] [CrossRef] [Green Version]
- Šimat, V.; Elabed, N.; Kulawik, P.; Ceylan, Z.; Jamroz, E.; Yazgan, H.; Čagalj, M.; Regenstein, J.M.; Özogul, F. Recent Advances in Marine-Based Nutraceuticals and Their Health Benefits. Mar. Drugs 2020, 18, 627. [Google Scholar] [CrossRef] [PubMed]
- Chiarelli, R.; Martino, C.; Agnello, M.; Bosco, L.; Roccheri, M.C. Autophagy as a defense strategy against stress: Focus on Paracentrotus lividus sea urchin embryos exposed to cadmium. Cell Stress Chaperones 2016, 21, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Liu, H.; Zhen, H.; Zhao, B. Autophagy and its role in regeneration and remodeling within invertebrate. Cell Biosci. 2020, 10, 111. [Google Scholar] [CrossRef]
- Van Soest, R.W.; Boury-Esnault, N.; Vacelet, J.; Dohrmann, M.; Erpenbeck, D.; De Voogd, N.J.; Santodomingo, N.; Vanhoorne, B.; Kelly, M.; Hooper, J.N. Global diversity of sponges (Porifera). PLoS ONE 2012, 7, e35105. [Google Scholar] [CrossRef]
- Invertebrates of the Coral Sea. Haliclona sp. Claudia Tay. 2012. Available online: https://www.gbri.org.au/Classes/Holothuriaatra%7CEmilyPurton/Haliclonasp%7CClaudiaTay.aspx (accessed on 23 March 2021).
- Yamazaki, H.; Wewengkang, D.S.; Kanno, S.; Ishikawa, M.; Rotinsulu, H.; Mangindaan, R.E.; Namikoshi, M. Papuamine and haliclonadiamine, obtained from an Indonesian sponge Haliclona sp., inhibited cell proliferation of human cancer cell lines. Nat. Prod. Res. 2013, 27, 1012–1015. [Google Scholar] [CrossRef]
- Kanno, S.; Yomogida, S.; Tomizawa, A.; Yamazaki, H.; Ukai, K.; Mangindaan, R.E.; Namikoshi, M.; Ishikawa, M. Papuamine causes autophagy following the reduction of cell survival through mitochondrial damage and JNK activation in MCF-7 human breast cancer cells. Int. J. Oncol. 2013, 43, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Killackey, S.A.; Philpott, D.J.; Girardin, S.E. Mitophagy pathways in health and disease. J. Cell Biol. 2020, 219, e202004029. [Google Scholar] [CrossRef]
- Kanno, S.I.; Yomogida, S.; Tomizawa, A.; Yamazaki, H.; Ukai, K.; Mangindaan, R.E.; Namikoshi, M.; Ishikawa, M. Combined effect of papuamine and doxorubicin in human breast cancer MCF-7 cells. Oncol. Lett. 2014, 8, 547–550. [Google Scholar] [CrossRef] [Green Version]
- Min, H.Y.; Jung, Y.; Park, K.H.; Lee, H.Y. Papuamine Inhibits Viability of Non-small Cell Lung Cancer Cells by Inducing Mitochondrial Dysfunction. Anti-cancer Res. 2020, 40, 323–333. [Google Scholar] [CrossRef]
- Haliclona caerulea (Hechtel, 1965) Blue Caribbean Sponge. Available online: https://www.sealifebase.ca/summary/Haliclona-caerulea.html# (accessed on 24 March 2021).
- Carneiro, R.F.; de Melo, A.A.; de Almeida, A.S.; Moura Rda, M.; Chaves, R.P.; de Sousa, B.L.; do Nascimento, K.S.; Sampaio, S.S.; Lima, J.P.; Cavada, B.S.; et al. H-3, a new lectin from the marine sponge Haliclona caerulea: Purification and mass spectrometric characterization. Int. J. Biochem. Cell Biol. 2013, 45, 2864–2873. [Google Scholar] [CrossRef]
- do Nascimento-Neto, L.G.; Cabral, M.G.; Carneiro, R.F.; Silva, Z.; Arruda, F.V.S.; Nagano, C.S.; Fernandes, A.R.; Sampaio, A.H.; Teixeira, E.H.; Videira, P.A. Halilectin-3, a Lectin from the Marine Sponge Haliclona caerulea, Induces Apoptosis and Autophagy in Human Breast Cancer MCF7 Cells Through Caspase-9 Pathway and LC3-II Protein Expression. Anti-cancer Agents Med. Chem. 2018, 18, 521–528. [Google Scholar] [CrossRef]
- Bijukumar, A.; Nair, A.S. (Eds.) Marine Biodiversity Informatics for Kerala. Available online: http://www.keralamarinelife.in (accessed on 25 March 2021).
- Keyzers, R.A.; Daoust, J.; Davies-Coleman, M.T.; Van Soest, R.; Balgi, A.; Donohue, E.; Roberge, M.; Andersen, R.J. Autophagy-modulating aminosteroids isolated from the sponge Cliona celata. Org. Lett. 2008, 10, 2959–2962. [Google Scholar] [CrossRef] [PubMed]
- Forestieri, R.; Donohue, E.; Balgi, A.; Roberge, M.; Andersen, R.J. Synthesis of clionamine B, an autophagy stimulating aminosteroid isolated from the sponge Cliona celata. Org. Lett. 2013, 15, 3918–3921. [Google Scholar] [CrossRef] [PubMed]
- Bergquist, P.R.; de Cook, S.C. Family Pseudoceratinidae Carter, 1885. In Systema Porifera: A Guide to the Classification of Sponges; Hooper, J.N.A., Van Soest, R.W.M., Eds.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2002; pp. 1086–1088. [Google Scholar]
- Kim, D.; Lee, I.S.; Jung, J.H.; Yang, S.I. Psammaplin A, a natural bromotyrosine derivative from a sponge, possesses the antibacterial activity against methicillin-resistant Staphylococcus aureus and the DNA gyrase-inhibitory activity. Arch. Pharm. Res. 1999, 22, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tabudravu, J.N.; Eijsink, V.G.; Gooday, G.W.; Jaspars, M.; Komander, D.; Legg, M.; Synstad, B.; van Aalten, D.M. Psammaplin A, a chitinase inhibitor isolated from the Fijian marine sponge Aplysinella rhax. Bioorg. Med. Chem. 2002, 10, 1123–1128. [Google Scholar] [CrossRef]
- Kim, D.H.; Shin, J.; Kwon, H.J. Psammaplin A is a natural prodrug that inhibits class I histone deacetylase. Exp. Mol. Med. 2007, 39, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Piña, I.C.; Gautschi, J.T.; Wang, G.Y.; Sanders, M.L.; Schmitz, F.J.; France, D.; Cornell-Kennon, S.; Sambucetti, L.C.; Remiszewski, S.W.; Perez, L.B.; et al. Psammaplins from the sponge Pseudoceratina purpurea: Inhibition of both histone deacetylase and DNA methyltransferase. J. Org. Chem. 2003, 68, 3866–3873. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, H.S.; Kang, Y.J.; Yoon, S.; Lee, J.; Choi, W.S.; Jung, J.H.; Kim, H.S. Psammaplin A induces Sirtuin 1-dependent autophagic cell death in doxorubicin-resistant MCF-7/adr human breast cancer cells and xenografts. Biochim. Biophys. Acta 2015, 1850, 401–410. [Google Scholar] [CrossRef]
- DeVorkin, L.; Choutka, C.; Gorski, S.M. The Interplay between Autophagy and Apoptosis. In Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging; Hayat, M.A., Ed.; Academic Press: Amsterdam, The Netherlands, 2014; pp. 369–383. [Google Scholar]
- Hu, W.; Chen, S.; Thorne, R.F.; Wu, M. TP53, TP53 Target Genes (DRAM, TIGAR), and Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 127–149. [Google Scholar] [CrossRef] [PubMed]
- Kreis, N.-N.; Louwen, F.; Yuan, J. The Multifaceted p21 (Cip1/Waf1/CDKN1A) in Cell Differentiation, Migration and Cancer Therapy. Cancers 2019, 11, 1220. [Google Scholar] [CrossRef] [Green Version]
- Richmond, M. (Ed.) A Guide to the Seashores of Eastern Africa and the Western Indian Ocean Islands; The Swedish International Development Co-operation: Stockholm, Sweden, 1997. [Google Scholar]
- Kobayashi, M.; Kawazoe, K.; Kitagawa, I. Araguspongines B, C, D, E, F, G, H, and J, new vasodilative bis-1-oxaquinolizidine alkaloids from an okinawan marine sponge, Xestospongia sp. Chem. Pharm. Bull. 1989, 37, 1676–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orabi, K.Y.; El Sayed, K.A.; Hamann, M.T.; Dunbar, D.C.; Al-Said, M.S.; Higa, T.; Kelly, M. Araguspongines K and L, new bioactive bis-1-oxaquinolizidine N-oxide alkaloids from Red Sea specimens of Xestospongia exigua. J. Nat. Prod. 2002, 65, 1782–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akl, M.R.; Ayoub, N.M.; Ebrahim, H.Y.; Mohyeldin, M.M.; Orabi, K.Y.; Foudah, A.I.; El Sayed, K.A. Araguspongine C induces autophagic death in breast cancer cells through suppression of c-Met and HER2 receptor tyrosine kinase signaling. Mar. Drugs 2015, 13, 288–311. [Google Scholar] [CrossRef] [Green Version]
- Ebrahim, H.Y.; El Sayed, K.A. Discovery of Novel Antiangiogenic Marine Natural Product Scaffolds. Mar. Drugs 2016, 14, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Tang, X.L.; Luo, X.C.; de Voog, N.J.; Li, P.L.; Li, G.Q. Aplysinopsin-type and Bromotyrosine-derived Alkaloids from the South China Sea Sponge Fascaplysinopsis reticulata. Sci. Rep. 2019, 9, 2248. [Google Scholar] [CrossRef] [Green Version]
- Segraves, N.L.; Robinson, S.J.; Garcia, D.; Said, S.A.; Fu, X.; Schmitz, F.J.; Pietraszkiewicz, H.; Valeriote, F.A.; Crews, P. Comparison of fascaplysin and related alkaloids: A study of structures, cytotoxicities, and sources. J. Nat. Prod. 2004, 67, 783–792. [Google Scholar] [CrossRef]
- Zheng, Y.L.; Lu, X.L.; Lin, J.; Chen, H.M.; Yan, X.J.; Wang, F.; Xu, W.F. Direct effects of fascaplysin on human umbilical vein endothelial cells attributing the anti-angiogenesis activity. Biomed. Pharmacother. 2010, 64, 527–533. [Google Scholar] [CrossRef]
- Chen, S.; Guan, X.; Wang, L.L.; Li, B.; Sang, X.B.; Liu, Y.; Zhao, Y. Fascaplysin inhibit ovarian cancer cell proliferation and metastasis through inhibiting CDK4. Gene 2017, 635, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Rath, B.; Hochmair, M.; Plangger, A.; Hamilton, G. Anticancer Activity of Fascaplysin against Lung Cancer Cell and Small Cell Lung Cancer Circulating Tumor Cell Lines. Mar. Drugs 2018, 16, 383. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Guru, S.K.; Manda, S.; Kumar, A.; Mintoo, M.J.; Prasad, V.D.; Sharma, P.R.; Mondhe, D.M.; Bharate, S.B.; Bhushan, S. A marine sponge alkaloid derivative 4-chloro fascaplysin inhibits tumor growth and VEGF mediated angiogenesis by disrupting PI3K/Akt/mTOR signaling cascade. Chem. Biol. Interact. 2017, 275, 47–60. [Google Scholar] [CrossRef]
- Dyshlovoy, S.A.; Fedorov, S.N.; Shubina, L.K.; Kuzmich, A.S.; Bokemeyer, C.; Keller-von Amsberg, G.; Honecker, F. Aaptamines from the marine sponge Aaptos sp. display anticancer activities in human cancer cell lines and modulate AP-1-, NF-κB-, and p53-dependent transcriptional activity in mouse JB6 Cl41 cells. Biomed. Res. Int. 2014, 2014, 469309. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.F.; Lee, M.G.; El-Shazly, M.; Lai, K.H.; Ke, S.C.; Su, C.W.; Shih, S.P.; Sung, P.J.; Hong, M.C.; Wen, Z.H.; et al. Isoaaptamine Induces T-47D Cells Apoptosis and Autophagy via Oxidative Stress. Mar. Drugs 2018, 16, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alderslade, P. Four new genera of soft corals (Coelenterata: Octocorallia), with notes on the classification of some established taxa. Zool. Med. Leiden. 2000, 74, 237–249. [Google Scholar]
- Weng, J.R.; Chiu, C.F.; Hu, J.L.; Feng, C.H.; Huang, C.Y.; Bai, L.Y.; Sheu, J.H. A Sterol from Soft Coral Induces Apoptosis and Autophagy in MCF-7 Breast Cancer Cells. Mar. Drugs 2018, 16, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotta-Loizou, I.; Giaginis, C.; Theocharis, S. The role of peroxisome proliferator-activated receptor-γ in breast cancer. Anticancer Agents Med. Chem. 2012, 12, 1025–1044. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.R.; Huang, C.Y.; Chen, B.W.; Tsai, Y.Y.; Shih, S.P.; Hwang, T.L.; Dai, C.F.; Wang, S.Y.; Sheu, J.H. New bioactive steroids from the soft coral Klyxum flaccidum. RSC Adv. 2015, 5, 12546–12554. [Google Scholar] [CrossRef]
- Matsubara, T.; Hayashi, A. Identification of molecular species of ceramide aminoethylphosphonate from oyster adductor by gas-liquid chromatography-mass spectrometry. Biochim. Biophys. Acta 1973, 296, 171–178. [Google Scholar]
- Matsubara, T. The structure and distribution of ceramide aminoethylphosphonates in the oyster (Ostrea gigas). Biochim. Biophys. Acta 1975, 388, 353–360. [Google Scholar] [PubMed]
- Moro, K.; Nagahashi, M.; Gabriel, E.; Takabe, K.; Wakai, T. Clinical application of ceramide in cancer treatment. Breast Cancer 2019, 26, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Chintalapati, M.; Truax, R.; Stout, R.; Portier, R.; Losso, J.N. In vitro and in vivo anti-angiogenic activities and inhibition of hormone-dependent and -independent breast cancer cells by ceramide methylaminoethylphosphonate. J. Agric. Food Chem. 2009, 57, 5201–5210. [Google Scholar] [CrossRef]
- Beg, A.A.; Ruben, S.M.; Scheinman, R.I.; Haskill, S.; Rosen, C.A.; Baldwin, A.S., Jr. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: A mechanism for cytoplasmic retention. Genes Dev. 1992, 6, 1899–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monkkonen, T.; Debnath, J. Inflammatory signaling cascades and autophagy in cancer. Autophagy 2018, 14, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Shen, H.-M.; Lim, L.H.K. The Role of Autophagy in Liver Cancer: Crosstalk in Signaling Pathways and Potential Therapeutic Targets. Pharmaceuticals 2020, 13, 432. [Google Scholar] [CrossRef]
- Kma, L.; Baruah, T.J. The interplay of ROS and the PI3K/Akt pathway in autophagy regulation. Biotechnol. Appl. Biochem. 2021. epub ahead of print. [Google Scholar] [CrossRef]
- Luparello, C.; Ragona, D.; Asaro, D.M.L.; Lazzara, V.; Affranchi, F.; Arizza, V.; Vazzana, M. Cell-Free Coelomic Fluid Extracts of the Sea Urchin Arbacia lixula Impair Mitochondrial Potential and Cell Cycle Distribution and Stimulate Reactive Oxygen Species Production and Autophagic Activity in Triple-Negative MDA-MB231 Breast Cancer Cells. J. Mar. Sci. Eng. 2020, 8, 261. [Google Scholar] [CrossRef] [Green Version]
- Vazzana, M.; Siragusa, T.; Arizza, V.; Buscaino, G.; Celi, M. Cellular responses and HSP70 expression during wound healing in Holothuria tubulosa (Gmelin, 1788). Fish Shellfish Immunol. 2015, 42, 306–315. [Google Scholar] [CrossRef]
- Mauro, M.; Queiroz, V.; Arizza, V.; Campobello, D.; Custódio, M.R.; Chiaramonte, M.; Vazzana, M. Humoral responses during wound healing in Holothuria tubulosa (Gmelin, 1788). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2021, 253, 110550. [Google Scholar] [CrossRef]
- Martino, C.; Bonaventura, R.; Byrne, M.; Roccheri, M.; Matranga, V. Effects of exposure to gadolinium on the development of geographically and phylogenetically distant sea urchins species. Mar. Environ. Res. 2017, 128, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.; Hidalgo, F.; García-Corcoles, M.T.; Ibáñez-Yuste, A.J.; Alonso, E.; Vilchez, J.L.; Zafra-Gómez, A. Bioaccumulation of perfluoroalkyl substances in marine echinoderms: Results of laboratory-scale experiments with Holothuria tubulosa Gmelin, 1791. Chemosphere 2019, 215, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Vazzana, M.; Mauro, M.; Ceraulo, M.; Dioguardi, M.; Papale, E.; Mazzola, S.; Arizza, V.; Beltrame, F.; Inguglia, L.; Buscaino, G. Underwater high frequency noise: Biological responses in sea urchin Arbacia lixula (Linnaeus, 1758). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2020, 242, 110650. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Ragona, D.; Asaro, D.M.L.; Lazzara, V.; Affranchi, F.; Celi, M.; Arizza, V.; Vazzana, M. Cytotoxic Potential of the Coelomic Fluid Extracted from the Sea Cucumber Holothuria tubulosa against Triple-Negative MDA-MB231 Breast Cancer Cells. Biology 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khotimchenko, Y. Pharmacological Potential of Sea Cucumbers. Int. J. Mol. Sci. 2018, 19, 1342. [Google Scholar] [CrossRef] [Green Version]
- Abdelkarem, F.M.; Desoky, E.K.; Nafady, A.M.; Allam, A.E.; Mahdy, A.; Ashour, A.; Shimizu, K. Diadema setosum: Isolation of bioactive secondary metabolites with cytotoxic activity toward human cervical cancer. Nat. Prod. Res. 2020, 4, 1–4. [Google Scholar] [CrossRef]
- Yurasakpong, L.; Apisawetakan, S.; Pranweerapaiboon, K.; Sobhon, P.; Chaithirayanon, K. Holothuria scabra Extract Induces Cell Apoptosis and Suppresses Warburg Effect by Down-Regulating Akt/mTOR/HIF-1 Axis in MDA-MB-231 Breast Cancer Cells. Nutr. Cancer 2020, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.S. Echinochrome Exhibits Antitumor Activity against Ehrlich Ascites Carcinoma in Swiss Albino Mice. Nutr. Cancer 2021, 73, 124–132. [Google Scholar] [CrossRef]
- Tasdemir, E.; Maiuri, M.C.; Tajeddine, N.; Vitale, I.; Criollo, A.; Vicencio, J.M.; Hickman, J.A.; Geneste, O.; Kroemer, G. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle 2007, 6, 2263–2267. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Wang, Y.; Wang, Z.; Wang, Y.P.; Zheng, H. Inhibiting autophagy increases epirubicin’s cytotoxicity in breast cancer cells. Cancer Sci. 2016, 107, 1610–1621. [Google Scholar] [CrossRef]
- Poole, L.P.; Macleod, K.F. Mitophagy in tumorigenesis and metastasis. Cell. Mol. Life Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer#:~:text=In%202020%2C%20there%20were%202.3,the%20world’s%20most%20prevalent%20cancer (accessed on 28 April 2021).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Barabadi, H.; Mahjoub, M.A.; Tajani, B.; Ahmadi, A.; Junejo, J.; Saravanan, M. Emerging Theranostic Biogenic Silver Nanomaterials for Breast Cancer: A Systematic Review. J. Cluster Sci. 2019, 30, 259–279. [Google Scholar] [CrossRef]
- Saravanan, M.; Vahidi, H.; Medina Cruz, D.; Vernet-Crua, A.; Mostafavi, E.; Stelmach, R.; Webster, T.J.; Mahjoub, M.A.; Rashedi, M.; Barabadi, H. Emerging Antineoplastic Biogenic Gold Nanomaterials for Breast Cancer Therapeutics: A Systematic Review. Int. J. Nanomed. 2020, 15, 3577–3595. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V. Autophagy in inflammation, infection, and immunometabolism. Immunity 2021, 54, 437–453. [Google Scholar] [CrossRef]
- Kouroumalis, E.; Voumvouraki, A.; Augoustaki, A.; Samonakis, D.N. Autophagy in liver diseases. World J. Hepatol. 2021, 13, 6–65. [Google Scholar] [CrossRef] [PubMed]
- Wargasetia, T.L.; Widodo, N. The Link of Marine Products with Autophagy-Associated Cell Death in Cancer Cell. Curr. Pharmacol. Rep. 2019, 5, 35–42. [Google Scholar] [CrossRef]





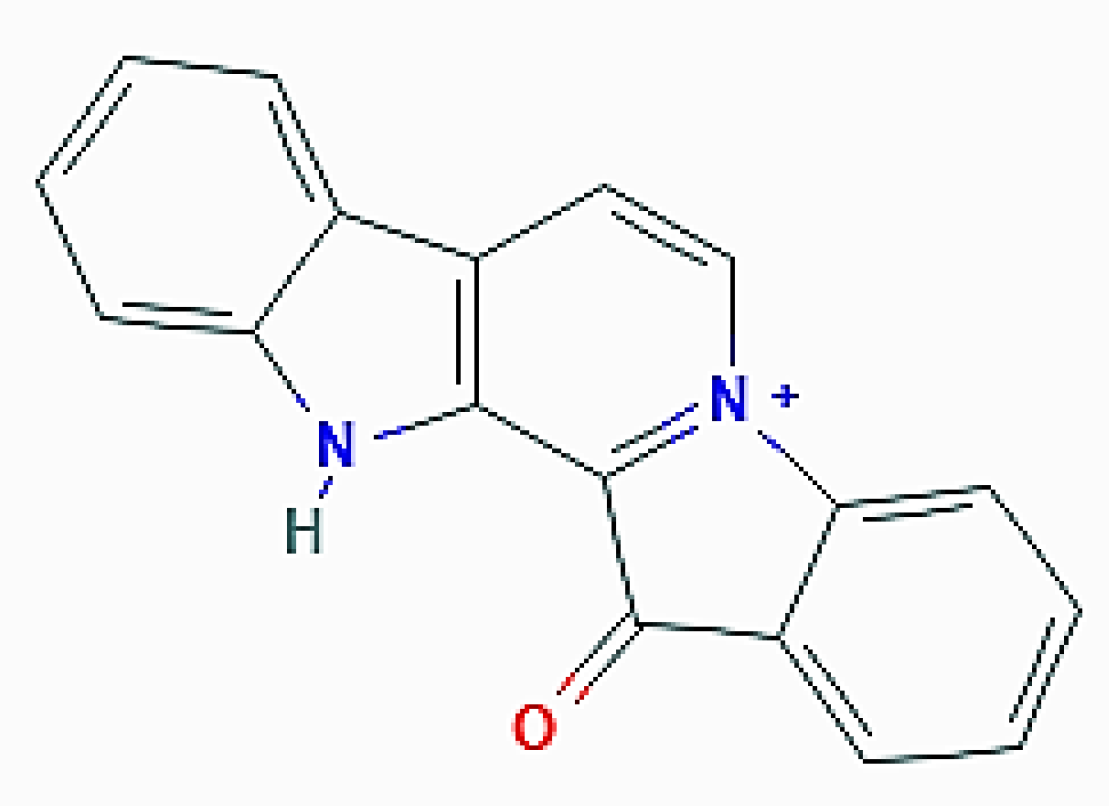

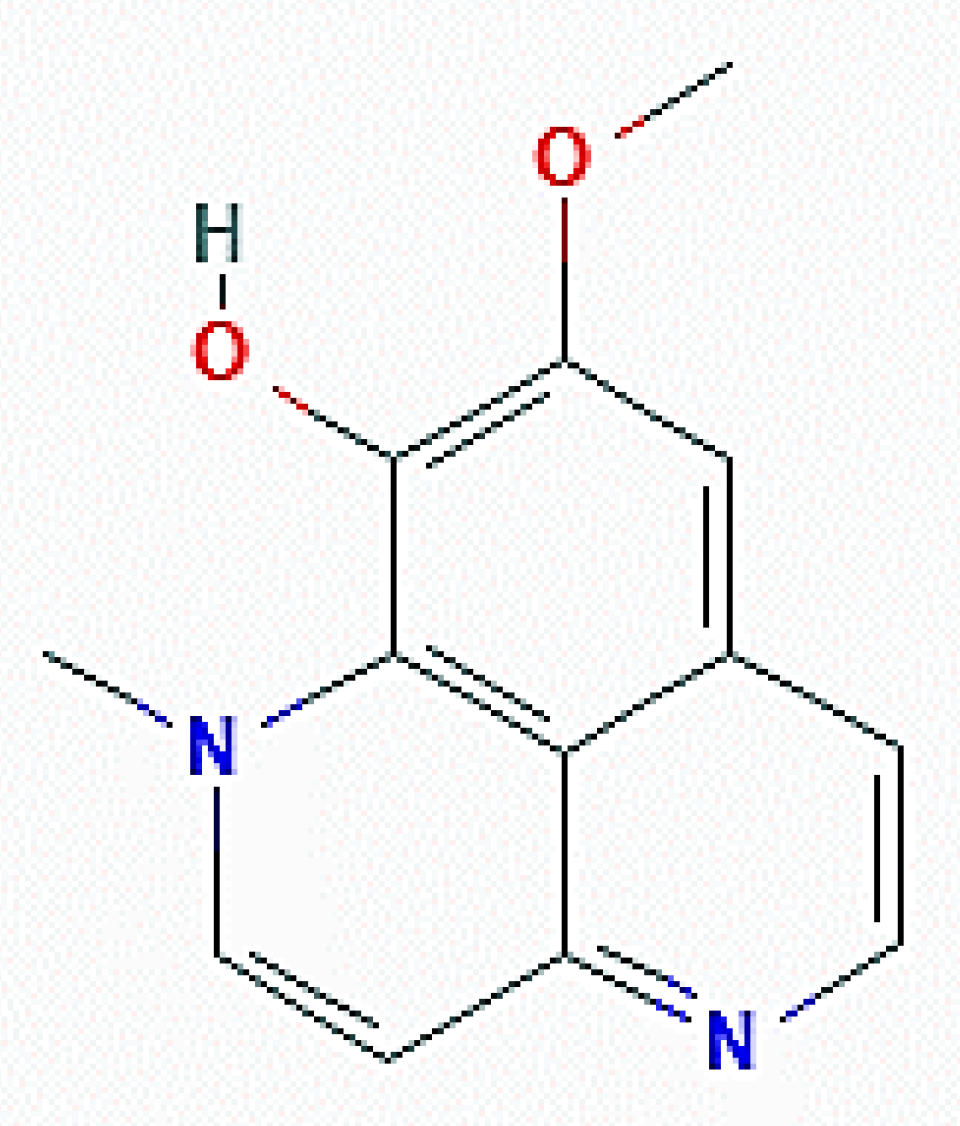





| Organism of Origin | Substance | Autophagy-Related Effects |
|---|---|---|
| Haliclona genus (Porifera) | Papuamine | Accumulation of total LC3 and LC3-II; possible mitophagy |
| H. caerulea (Porifera) | Halilectin-3 | Upregulation of MAP1LC3B gene expression; accumulation of LC3-II |
| C. celata (Porifera) | Clionamine A-D | increase of cytoplasmic green fluorescent protein (GFP)-LC3 puncta |
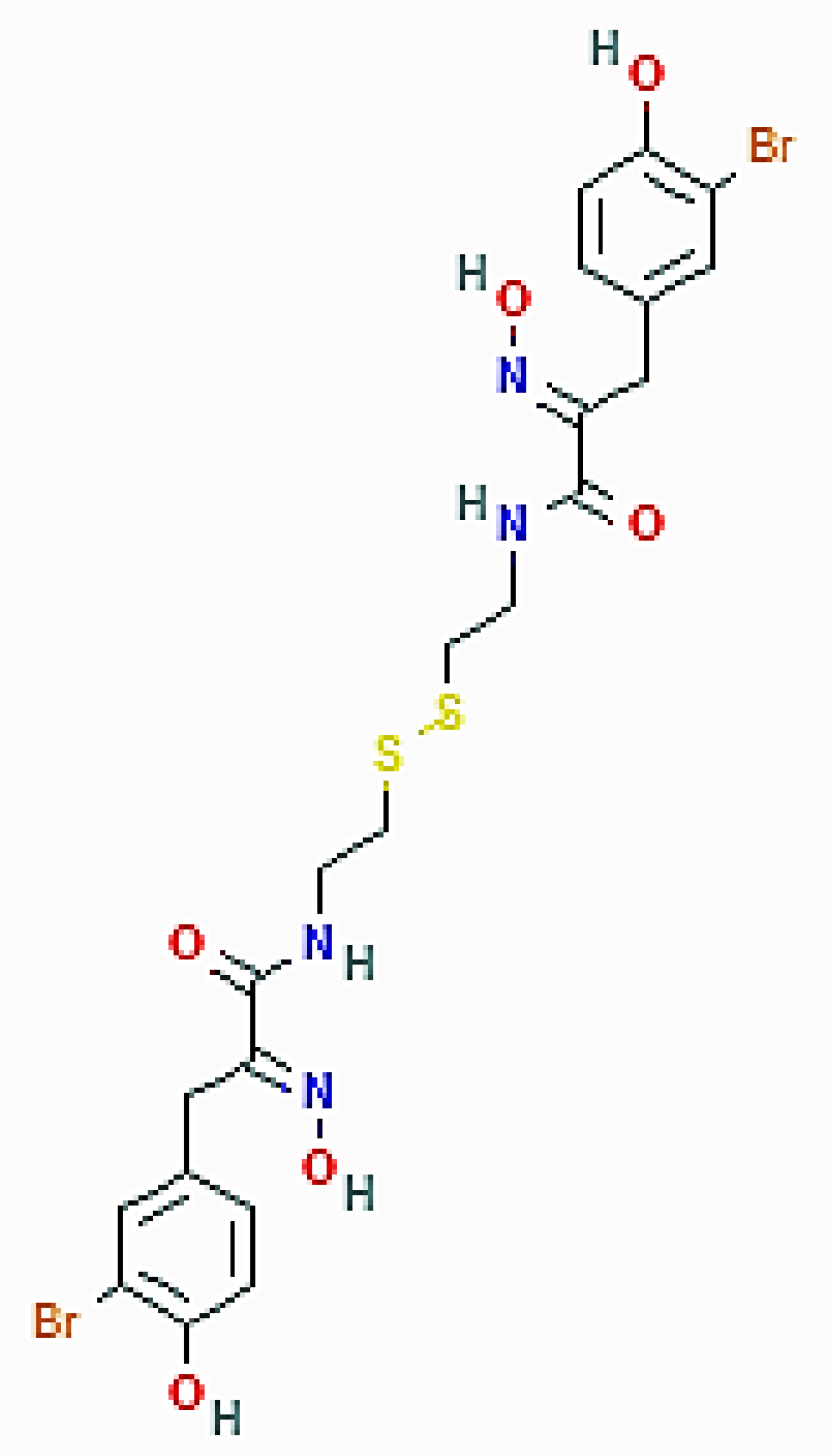
| Psammaphysilla genus (Porifera) | Psammaplin A | Increase of acidic vacuolar organelles; upregulation of LC3, ATG-3, -5, -7, -12, and beclin-1; downregulation of p62/SQSTM1 protein |
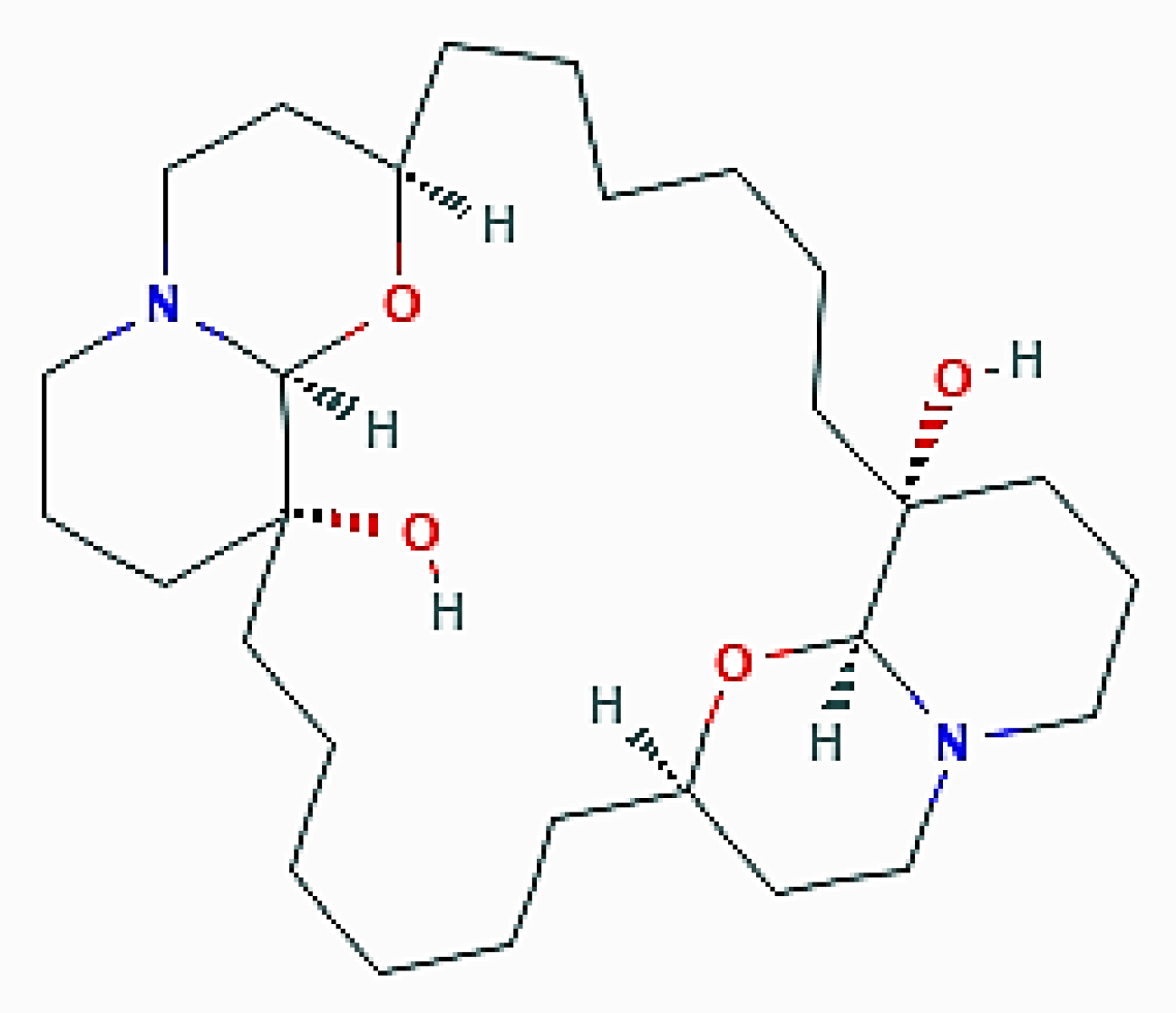
| X. esigua (Porifera) | Araguspongine C | Increase of fluorescent autophagosomes; upregulation of total LC3, Beclin-1, ATG-5, -7, and -16L1 |
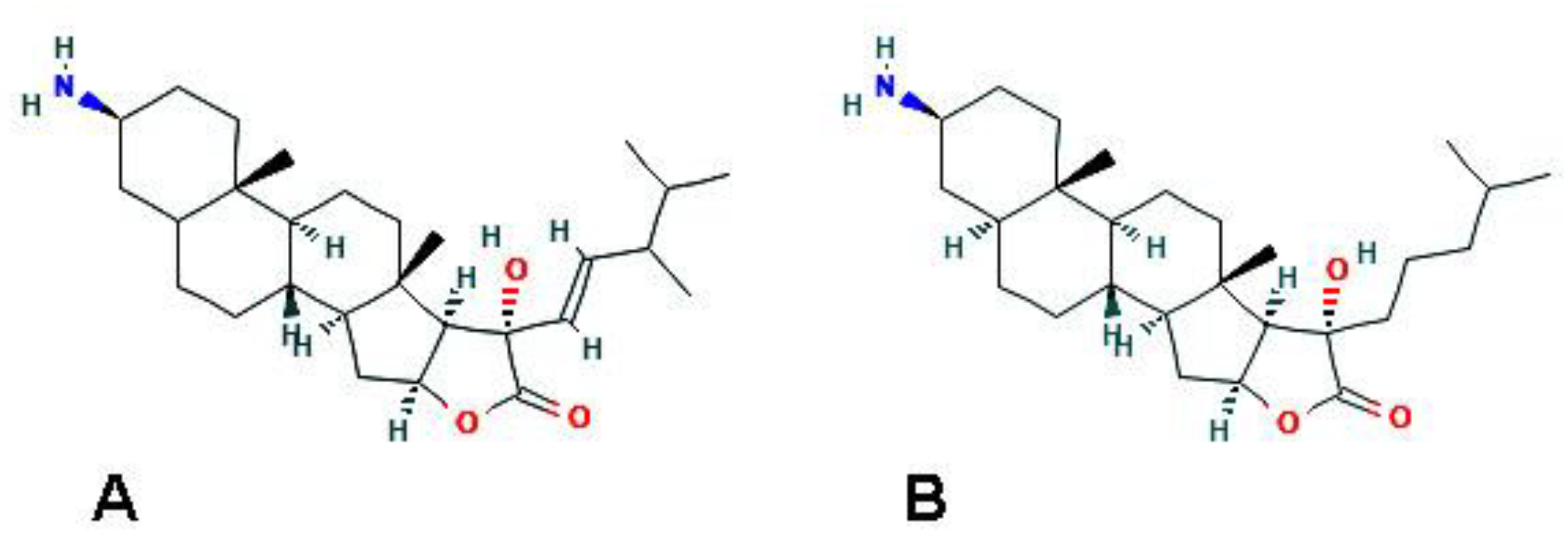
| F. reticulata (Porifera) | Fascaplysin | Increase of acidic vacuolar organelles; upregulation of LC3-II (by 4-chloro-fascaplysin derivative) |
| Aaptos genus (Porifera) | Isoaaptamine | Increase of acridine orange-positive vacuoles; upregulation of LC3-II and p62/SQSTM1; downregulation of mTOR |
| K. flaccidum (Cnidaria) | 3β,11-dihydroxy-9,11- secogorgost-5-en-9-one | Increase of acidic vesicular organelles; upregulation of LC3-II and p62 |
| C. virginica (Mollusca) | N-methylaminoethyl phosphonate | Increase of fluorescent autophagosomes; upregulation of beclin-1 |
| A. lixula (Echinodermata) | Cell-free aqueous extracts of the coelomic fluid | Increase of acidic vesicular organelles; upregulation of beclin-1, LC3-II, and total LC3 |
| H. tubulosa (Echinodermata) | Cell-free aqueous extracts of the coelomic fluid | Increase of acidic vesicular organelles; upregulation of beclin-1, LC3-II, and total LC3; possible mitophagy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luparello, C. Marine Animal-Derived Compounds and Autophagy Modulation in Breast Cancer Cells. Foundations 2021, 1, 3-20. https://0-doi-org.brum.beds.ac.uk/10.3390/foundations1010002
Luparello C. Marine Animal-Derived Compounds and Autophagy Modulation in Breast Cancer Cells. Foundations. 2021; 1(1):3-20. https://0-doi-org.brum.beds.ac.uk/10.3390/foundations1010002
Chicago/Turabian StyleLuparello, Claudio. 2021. "Marine Animal-Derived Compounds and Autophagy Modulation in Breast Cancer Cells" Foundations 1, no. 1: 3-20. https://0-doi-org.brum.beds.ac.uk/10.3390/foundations1010002






