SARS–CoV–2 and Food—How Confident Are We about Them?
Abstract
:1. Introduction
- Can SARS–CoV–2 infect food animals? Can the virus multiply in a food animal and be detected in food products of animal origin?
- Can SARS–CoV–2 be found in water and retain its infectivity for an adequate time?
- Can food get contaminated by SARS–CoV–2 through its various production processes?
- Can SARS–CoV–2 be transmitted through water or food, and if yes, which are the possible transmission pathways related to water and food ingestion?
2. Human CoVs
3. Zoonotic Potential of Human CoVs
4. Transmission Pathways Related to Ingestion
5. Occurrence and Survival in Water
6. Food and CoVs
6.1. Other CoVs
6.2. Is It Foodborne?
6.3. Surfaces
6.4. CoV Behaviour in Food Treatments
6.5. Food Type and SARS–CoV–2
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. [Google Scholar] [CrossRef] [PubMed]
- Goraichuk, I.V.; Arefiev, V.; Stegniy, B.T.; Gerilovych, A.P. Zoonotic and Reverse Zoonotic Transmissibility of SARS-CoV-2. Virus Res. 2021, 302, 198473. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Kingsley, D.H. Emerging foodborne and agriculture related viruses. Microbiol. Spectrum 2016, 4. [Google Scholar] [CrossRef]
- Jalava, K. First respiratory transmitted food borne outbreak? Int. J. Hyg. Environ. Health 2020, 226, 113490. [Google Scholar] [CrossRef]
- Heller, L.; Mota, C.R.; Greco, D.B. COVID-19 faecal-oral transmission: Are we asking the right questions? Sci. Tot. Environ. 2020, 729, 138919. [Google Scholar] [CrossRef]
- Bondad-Reantaso, M.G.; Mackinnon, B.; Bin, H.; Jie, H.; Tang-Nelson, K.; Surachetpong, W.; Alday-Sanz, V.; Salman, M.; Brun, E.; Karunasagar, I.; et al. Viewpoint: Sars-CoV-2 (the cause of COVID-19 in humans) is not known to infect aquatic food animals nor contaminate their products. Asian Fish. Sci. 2020, 33, 74–78. [Google Scholar] [CrossRef]
- Carducci, A.; Federigi, I.; Liu, D.; Thompson, J.R.; Verani, M. Making Waves: Coronavirus detection, presence and persistence in the water environment: State of the art and knowledge needs for public health. Water Res. 2020, 179, 115907. [Google Scholar] [CrossRef]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods—A scoping review. Water Res. 2020, 179, 115899. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Alamin, M.; Kuroda, K.; Dhangar, K.; Hata, A.; Yamaguchi, H.; Honda, R. Potential discharge, attenuation and exposure risk of SARS-CoV-2 in natural water bodies receiving treated wastewater. NPJ Clean Water 2021, 4, 8. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/CoVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef] [PubMed]
- Petala, M.; Dafou, D.; Kostoglou, M.; Karapantsios, T.; Kanata, E.; Chatziefstathiou, A.; Kotoulas, K.; Arsenakis, M.; Roilides, E.; Sklaviadis, T.; et al. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. Case study: The city of Thessaloniki in Greece. Sci. Total Environ. 2021, 755 Pt 1, 142855. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, Z.; Meral, R.; Cetinkaya, T. Relevance of SARS-CoV-2 in food safety and food hygiene: Potential preventive measures, suggestions and nanotechnological approaches. Virus Dis. 2020, 31, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhary, J.; Dheeman, S.; Sharma, V.; Katiyar, P.; Karn, S.K.; Sarangi, M.K.; Chauhan, A.K.; Chauhan, A.K.; Verma, G.; Baliyan, N. Insights of severe acute respiratory syndrome coronavirus (SARS-CoV-2) pandemic: A current review. Biol. Proc. 2021, 23, 5. [Google Scholar] [CrossRef]
- Kapikian, A.Z. The coronaviruses. Dev. Biol. Stand. 1975, 28, 42–64. [Google Scholar]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Van Der Hoek, L. Human coronaviruses: What do they cause? Antiviral Ther. 2007, 12, 651–658. [Google Scholar]
- Tyrrell, D.A.; Cohen, S.; Schlarb, J.E. Signs and symptoms in common colds. Epidemiol. Infect. 1993, 111, 143–156. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef]
- Stout, A.E.; Millet, J.K.; Stanhope, M.J.; Whittaker, G.R. Furin cleavage sites in the spike proteins of bat and rodent coronaviruses: Implications for virus evolution and zoonotic transfer from rodent species. One Health 2021, 13, 100282. [Google Scholar] [CrossRef] [PubMed]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Cheng, V.C.; Lau, S.K.; Woo, P.C.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007, 20, 660–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilgenfeld, R.; Peiris, M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013, 100, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wang, B.; Yuan, T.; Chen, X.; Ao, Y.; Fitzpatrick, T.; Li, P.; Zhou, Y.; Lin, Y.F.; Duan, Q.; et al. Clinical characteristics of coronavirus disease 2019 (CoVID-19) in China: A systematic review and meta-analysis. J. Infect. 2020, 80, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Parasa, S.; Desai, M.; Thoguluva Chandrasekar, V.; Patel, H.K.; Kennedy, K.F.; Roesch, T.; Spadaccini, M.; Colombo, M.; Gabbiadini, R.; Artifon, E.L.A.; et al. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with Coronavirus Disease 2019: A systematic review and meta-analysis. JAMA Netw. Open 2020, 3, e2011335. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.S.; Ye, Z.W.; Fung, S.Y.; Chan, C.P.; Jin, D.Y. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Bande, F.; Arshad, S.S.; Omar, A.R.; Bejo, M.H.; Abubakar, M.S.; Abba, Y. Pathogenesis and diagnostic approaches of avian infectious bronchitis. Adv. Virol. 2016, 2016, 4621659. [Google Scholar] [CrossRef] [Green Version]
- Bonilauri, P.; Rugna, G. Animal Coronaviruses and SARS-COV-2 in Animals, What Do We Actually Know? Life 2021, 11, 123. [Google Scholar] [CrossRef]
- Erles, K.; Brownlie, J. Canine respiratory coronavirus: An emerging pathogen in the canine infectious respiratory disease complex. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 815–825. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Broderson, J.R.; Murphy, F.A. New strain of mouse hepatitis virus as the cause of lethal enteritis in infant mice. Infect. Immun. 1979, 24, 508–522. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Pusterla, N.; Vin, R.; Leutenegger, C.M.; Mittel, L.D.; Divers, T.J. Enteric coronavirus infection in adult horses. Vet. J. 2018, 231, 13–18. [Google Scholar] [CrossRef]
- Decaro, N.; Buonavoglia, C. Canine coronavirus: Not only an enteric pathogen. Vet. Clin. N. Am. Small Anim. Pract. 2011, 41, 1121–1132. [Google Scholar] [CrossRef]
- Mahdy, M.A.A.; Younis, W.; Ewaida, Z. An Overview of SARS-CoV-2 and Animal Infection. Front. Vet. Sci. 2020, 7, 596391. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [Green Version]
- Benvenuto, D.; Giovanetti, M.; Ciccozzi, A.; Spoto, S.; Angeletti, S.; Ciccozzi, M. The 2019-new coronavirus epidemic: Evidence for virus evolution. J. Med. Virol. 2020, 92, 455–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Food and Agriculture Organization of the United Nations; World Health Organization. Viruses in Food: Scientific Advice to Support Risk Management Activities: Meeting Report; Lawrence, T., Ed.; Microbiological Risk Assessment Series No. 13; FAO: Rome, Italy, 2008; Available online: http://www.fao.org/3/i0451e/i0451e.pdf (accessed on 27 September 2021).
- Coleman, C.M.; Frieman, M.B. Emergence of the Middle East Respiratory Syndrome Coronavirus. PLoS Pathog. 2013, 9, e1003595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- To, K.K.; Hung, I.F.; Chan, J.F.; Yuen, K.Y. From SARS coronavirus to novel animal and human coronaviruses. J. Thorac. Dis. 2020, 5 (Suppl. S2), S103–S108. [Google Scholar] [CrossRef]
- Gao, H.; Yao, H.; Yang, S.; Li, L. From SARS to MERS: Evidence and speculation. Front. Med. 2020, 10, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Dhama, K.; Pawde, A.M.; Gortázar, C.; Tiwari, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; de la Fuente, J.; Michalak, I.; Attia, Y.A. SARS-CoV-2 in animals: Potential for unknown reservoir hosts and public health implications. Vet. Q. 2021, 41, 181–201. [Google Scholar] [CrossRef]
- Sun, K.; Gu, L.; Ma, L.; Duan, Y. Atlas of ACE2 gene expression reveals novel insights into transmission of SARS-CoV-2. Heliyon 2021, 7, e05850. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Meekins, D.A.; Morozov, I.; Trujillo, J.D.; Gaudreault, N.N.; Bold, D.; Carossino, M.; Artiaga, B.L.; Indran, S.V.; Kwon, T.; Balaraman, V.; et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 2278–2288. [Google Scholar] [CrossRef]
- Godoy, M.G.; Kibenge, M.J.T.; Kibenge, F.S.B. SARS-CoV-2 transmission via aquatic food animal species or their products: A review. Aquaculture 2021, 536, 736460. [Google Scholar] [CrossRef]
- Pickering, B.S.; Smith, G.; Pinette, M.M.; Embury-Hyatt, C.; Moffat, E.; Marszal, P.; Lewis, C.E. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome Coronavirus 2. Emerg. Infect. Dis. 2021, 27, 104–112. [Google Scholar] [CrossRef]
- Ulrich, L.; Wernike, K.; Hoffmann, D.; Mettenleiter, T.C.; Beer, M. Experimental infection of cattle with SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2979–2981. [Google Scholar] [CrossRef] [PubMed]
- Di Teodoro, G.; Valleriani, F.; Puglia, I.; Monaco, F.; Di Pancrazio, C.; Luciani, M.; Krasteva, I.; Petrini, A.; Marcacci, M.; D’Alterio, N.; et al. SARS-CoV-2 replicates in respiratory ex vivo organ cultures of domestic ruminant species. Vet. Microbiol. 2021, 252, 108933. [Google Scholar] [CrossRef]
- Deng, J.; Jin, Y.; Liu, Y.; Sun, J.; Hao, L.; Bai, J.; Huang, T.; Lin, D.; Jin, Y.; Tian, K. Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound. Emerg. Dis. 2020, 67, 1745–1749. [Google Scholar] [CrossRef] [PubMed]
- Hoseinzadeh, E.; Javan, S.; Farzadkia, M.; Mohammadi, F.; Hossini, H.; Taghavi, M. An updated min-review on environmental route of the SARS-CoV-2 transmission. Ecotoxicol. Environ. Saf. 2020, 202, 111015. [Google Scholar] [CrossRef]
- La Rosa, G.; Fratini, M.; Della Libera, S.; Iaconelli, M.; Muscillo, M. Viral infections acquired indoors through airborne, droplet or contact transmission. Ann. Ist. Super. Sanita 2013, 49, 124–132. [Google Scholar] [CrossRef]
- Khan, S.; Siddique, R.; Shereen, M.A.; Ali, A.; Liu, J.; Bai, Q.; Bashir, N.; Xue, M. Emergence of a Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2: Biology and Therapeutic Options. J. Clin. Microbiol. 2020, 58, e00187-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, A.; Gkogka, E.; Le Guyader, F.S.; Loisy-Hamon, F.; Lee, A.; van Lieshout, L.; Marthi, B.; Myrmel, M.; Sansom, A.; Schultz, A.C.; et al. Foodborne viruses: Detection, risk assessment, and control options in food processing. Int. J. Food Microbiol. 2018, 285, 110–128. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y.; et al. SARS-CoV-2 Infection in Children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Zhang, X.; Xu, H. Don’t Overlook Digestive Symptoms in Patients With 2019 Novel Coronavirus Disease (COVID-19). Clin. Gastroenterol. Hepatol. 2020, 18, 1636–1637. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heneghan, C.J.; Spencer, E.A.; Brassey, J.; Plüddemann, A.; Onakpoya, I.; Evans, D.H.; Conly, J.; Jefferson, T. SARS-CoV-2 and the role of orofecal transmission: A systematic review. F1000Research 2021, 10, 231. [Google Scholar] [CrossRef]
- Zhang, J.; Garrett, S.; Sun, J. Gastrointestinal symptoms, pathophysiology, and treatment in COVID-19. Genes Dis. 2021, 8, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Venutolo, G.; Pujolassos Tanyà, M.; Delli Carri, M.; Landolfi, A.; Fasano, A. COVID-19 and the gastrointestinal tract: Source of infection or merely a target of the inflammatory process following SARS-CoV-2 infection? World J. Gastroenterol. 2021, 27, 1406–1418. [Google Scholar] [CrossRef]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833. [Google Scholar] [CrossRef]
- Leung, W.K.; To, K.F.; Chan, P.K.; Chan, H.L.; Wu, A.K.; Lee, N.; Yuen, K.Y.; Sung, J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 2003, 125, 1011–1017. [Google Scholar] [CrossRef] [Green Version]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Jones, T.C.; Vollmar, P.; Rothe, C.; Hoelscher, M.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Xiao, Q.; Tang, Y.; Qu, X.; Kuang, L.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Xing, Y.H.; Ni, W.; Wu, Q.; Li, W.J.; Li, G.J.; Wang, W.D.; Tong, J.N.; Song, X.F.; Wing-Kin Wong, G.; Xing, Q.S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020, 53, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Blondin-Brosseau, M.; Harlow, J.; Doctor, T.; Nasheri, N. Examining the persistence of human Coronavirus 229E on fresh produce. Food Microbiol. 2021, 98, 103780. [Google Scholar] [CrossRef]
- Giacobbo, A.; Rodrigues, M.A.S.; Zoppas Ferreira, J.; Bernardes, A.M.; de Pinho, M.N. A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Sci. Total. Environ. 2021, 774, 145721. [Google Scholar] [CrossRef]
- O’Brien, B.; Goodridge, L.; Ronholm, J.; Nasheri, N. Exploring the potential of foodborne transmission of respiratory viruses. Food Microbiol. 2021, 95, 103709. [Google Scholar] [CrossRef]
- Mallapaty, S. How sewage could reveal true scale of coronavirus outbreak. Nature 2020, 580, 176–177. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.W.; Li, J.S.; Jin, M.; Zhen, B.; Kong, Q.X.; Song, N.; Xiao, W.J.; Yin, J.; Wei, W.; Wang, G.J.; et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 126, 171–177. [Google Scholar] [CrossRef]
- Corsi, S.R.; Borchardt, M.A.; Spencer, S.K.; Hughes, P.E.; Baldwin, A.K. Human and bovine viruses in the Milwaukee River watershed: Hydrologically relevant representation and relations with environmental variables. Sci. Total Environ. 2014, 490, 849–860. [Google Scholar] [CrossRef] [Green Version]
- Blanco, A.; Abid, I.; Al-Otaibi, N.; Perez-Rodríguez, F.J.; Fuentes, C.; Guix, S.; Pintó, R.M.; Boschm, A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019, 11, 184e192. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of CoVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.M.; Moulin, L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv 2020. [Google Scholar] [CrossRef]
- Bandala, E.R.; Kruger, B.R.; Cesarino, I.; Leao, A.L.; Wijesiri, B.; Goonetilleke, A. Impacts of COVID-19 pandemic on the wastewater pathway into surface water: A review. Sci. Total Environ. 2021, 774, 145586. [Google Scholar] [CrossRef]
- Center for Disease Control and Infection. Food and Coronavirus Disease 2019 (COVID-19). 2020. Available online: https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/food-and-COVID-19.html (accessed on 15 July 2021).
- Sala-Comorera, L.; Reynolds, L.J.; Martin, N.A.; O’Sullivan, J.J.; Meijer, W.G.; Fletcher, N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021, 29, 117090. [Google Scholar] [CrossRef]
- Gundy, P.; Gerba, C.; Pepper, I. Survival of Coronaviruses in Water and Wastewater. Food Environ. Virol. 2009, 1, 10–14. [Google Scholar] [CrossRef] [Green Version]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- De Oliveira, L.C.; Torres-Franco, A.F.; Lopes, B.C.; Santos, B.S.Á.D.S.; Costa, E.A.; Costa, M.S.; Reis, M.T.P.; Melo, M.C.; Polizzi, R.B.; Teixeira, M.M.; et al. Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 2021, 195, 117002. [Google Scholar] [CrossRef]
- World Health Organization; United Nations Children’s Fund (UNICEF). Water, Sanitation, Hygiene, and Waste Management for the COVID-19 Virus: Interim Guidance, 19 March 2020; World Health Organization: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/handle/10665/331499 (accessed on 15 July 2021).
- Geller, C.; Varbanov, M.; Duval, R.E. Human coronaviruses: Insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses 2012, 4, 3044–3068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organisation. WHO Guidelines for the Global Surveillance of Severe Acute Respiratory Syndrome (SARS); WHO: Geneva, Switzerland, 2004; Available online: https://www.who.int/csr/resources/publications/WHO_CDS_CSR_ARO_2004_1.pdf?ua=1 (accessed on 15 July 2021).
- Todd, E.; Grieg, J. Viruses of foodborne origin: A review. Virus Adapt. Treat. 2015, 7, 25–45. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, C.; Zhao, G.; Chu, H.; Wang, D.; Yan, H.H.-N.; Poon, V.K.; Wen, L.; Wong, B.H.; Zhao, X.; et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017, 3, eaao4966. [Google Scholar] [CrossRef] [Green Version]
- Duda-Chodak, A.; Lukasiewicz, M.; Zięć, G.; Florkiewicz, A.; Filipiak-Florkiewicz, A. COVID-19 pandemic and food: Present knowledge, risks, consumers fears and safety. Trends Food Sci. Technol. 2020, 105, 145–160. [Google Scholar] [CrossRef]
- Hartman, A.L.; Nambulli, S.; McMillen, C.M.; White, A.G.; Tilston-Lunel, N.L.; Albe, J.R.; Cottle, E.; Dunn, M.D.; Frye, L.J.; Gilliland, T.H.; et al. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS Pathog. 2020, 16, e1008903. [Google Scholar] [CrossRef]
- Lee, A.C.; Zhang, A.J.; Chan, J.F.; Li, C.; Fan, Z.; Liu, F.; Chenm, Y.; Liangm, R.; Sridhar, S.; Cai, J.P.; et al. Oral SARS-CoV-2 Inoculation Establishes Subclinical Respiratory Infection with Virus Shedding in Golden Syrian Hamsters. Cell Rep. Med. 2020, 1, 100121. [Google Scholar] [CrossRef]
- Todd, E.C.; Greig, J.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 6. Transmission and survival of pathogens in the food processing and preparation environment. J. Food Prot. 2009, 72, 202–219. [Google Scholar] [CrossRef]
- Bachofen, C. Selected Viruses Detected on and in our Food. Curr. Clin. Micro. Rpt. 2018, 5, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Nasheri, N.; Vester, A.; Petronella, N. Foodborne viral outbreaks associated with frozen produce. Epidemiol. Infect. 2019, 147, E291. [Google Scholar] [CrossRef] [PubMed]
- Rönnqvist, M.; Maunula, L. Noroviruses on surfaces: Detection, persistence, disinfection and role in environmental transmission. Future Virol. 2016, 11, 207–217. [Google Scholar] [CrossRef]
- Lodder, W.; de Roda Husman, A. SARS-CoV-2 in wastewater: Potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020, 1253, 533–534. [Google Scholar] [CrossRef]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Gao, J.; Yao, L.; Zeng, L.; Liu, Q.; Zhou, Q.; Zhang, H.; Lu, D.; Fu, J.; Liu, Q.S.; et al. Evidence of foodborne transmission of the coronavirus (COVID-19) through the animal products food supply chain. Environ. Sci. Technol. 2021, 55, 2713–2716. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-D.; Wang, Z.-Y.; Zhang, S.-F.; Li, X.; Li, L.; Li, C.; Cui, Y.; Fu, R.B.; Dong, Y.Z.; Chi, X.Y.; et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital Wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020, 26, 1583–1591. [Google Scholar] [CrossRef]
- McDermott, C.V.; Alicic, R.Z.; Harden, N.; Cox, E.J.; Scanlan, J.M. Put a lid on it: Are faecal bio-aerosols a route of transmission for SARS-CoV-2? J. Hosp. Infect. 2020, 105, 397–398. [Google Scholar] [CrossRef]
- Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Wałecka-Zacharska, E.; Kwiecińska-Piróg, J.; Radtke, L.; Gospodarek-Komkowska, E.; Skowron, K. SARS-CoV-2 in the environment-Non-droplet spreading routes. Sci. Total Environ. 2021, 770, 145260. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.-L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Kasloff, S.B.; Leung, A.; Strong, J.E.; Funk, D.; Cutts, T. Stability of SARS-CoV-2 on critical personal protective equipment. Sci. Rep. 2021, 11, 984. [Google Scholar] [CrossRef]
- Sizun, J.; Yu, M.W.; Talbot, P. Survival of human coronaviruses 229E and OC43 in suspension and after drying on surfaces: A possible source of hospital-acquired infections. J. Hosp. Infect. 2000, 46, 55–60. [Google Scholar] [CrossRef]
- Duan, S.M.; Zhao, X.S.; Wen, R.F.; Huang, J.J.; Pi, G.H.; Zhang, S.X.; Han, J.; Bi, S.L.; Ruan, L.; Dong, X.P. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed. Environ. Sci. 2003, 16, 246–255. [Google Scholar]
- Lai, M.Y.; Cheng, P.K.; Lim, W.W. Survival of severe acute respiratory syndrome coronavirus. Clin. Infect. Dis. 2005, 41, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Hirose, R.; Ikegaya, H.; Naito, Y.; Watanabe, N.; Yoshida, T.; Bandou, R.; Daidoji, T.; Itoh, Y.; Nakaya, T. Survival of SARS-CoV-2 and influenza virus on the human skin: Importance of hand hygiene in COVID-19. Clin. Infect. Dis. 2020, ciaa1517. [Google Scholar] [CrossRef] [PubMed]
- Riddell, S.; Goldie, S.; Hill, A.; Eagles, D.; Drew, T. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol. J. 2020, 17, 145. [Google Scholar] [CrossRef] [PubMed]
- Rabenau, H.F.; Cinatl, J.; Morgenstern, B.; Bauer, G.; Preiser, W.; Doerr, H.W. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2005, 194, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.H.; Peiris, J.S.; Lam, S.Y.; Poon, L.L.; Yuen, K.Y.; Seto, W.H. The Effects of Temperature and Relative Humidity on the Viability of the SARS Coronavirus. Adv. Virol. 2011, 2011, 734690. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Munster, V.J. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013, 18, 20590. [Google Scholar] [CrossRef] [Green Version]
- Warnes, S.L.; Little, Z.R.; Keevil, C.W. Human coronavirus 229E remains infectious on common touch surface materials. MBio 2015, 6, e01697-15. [Google Scholar] [CrossRef] [Green Version]
- Pastorino, B.; Touret, F.; Gilles, M.; de Lamballerie, X.; Charrel, R.N. Prolonged Infectivity of SARS-CoV-2 in Fomites. Emerg. Infect. Dis. 2020, 26, 2256–2257. [Google Scholar] [CrossRef]
- Liu, Y.; Li, T.; Deng, Y.; Liu, S.; Zhang, D.; Li, H.; Wang, X.; Jia, L.; Han, J.; Bei, Z.; et al. Stability of SARS-CoV-2 on environmental surfaces and in human excreta. J. Hosp. Infect. 2021, 107, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Thippareddi, H.; Balamurugan, S.; Patel, J.; Singh, M.; Brassard, J. Coronaviruses—Potential human threat from foodborne transmission? Lebensm. Wiss. Technol. 2020, 31, 110147. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Li, J.; Guo, T.; Zhen, B.; Kong, Q.; Yi, B.; Li, Z.; Song, N.; Jin, M.; Xiao, W.; et al. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci. Technol. 2005, 52, 213–221. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand-Moreau, Q.A.A.; Mackenzie, G.; Bowley, J.; Straube, S.; Chan, X.H.; Zelyas, N.; Greenhalgh, T. COVID-19 in Meat and Poultry Facilities: A Rapid Review and Lay Media Analysis. 2020. Available online: https://www.cebm.net/covid-19/what-explains-the-high-rate-of-sars-cov-2-transmission-in-meat-and-poultry-facilities-2/ (accessed on 15 July 2021).
- Ursachi, C.Ș.; Munteanu, F.; Cioca, G. The safety of slaughterhouse workers during the pandemic crisis. Int. J. Environ. Res. Public Health 2021, 18, 2633. [Google Scholar] [CrossRef] [PubMed]
- Bundesinstitut für Risikobewertung (BfR). Can the New Type of Coronavirus Be Transmitted via Food and Objects? Available online: https://www.bfr.bund.de/cm/349/can-the-new-type-of-coronavirus-be-transmitted-via-food-and-objects.pdf (accessed on 15 July 2021).
- Galanakis, C.M. The Food Systems in the Era of the Coronavirus (COVID-19) Pandemic Crisis. Foods 2020, 9, 523. [Google Scholar] [CrossRef]
- Yépiz-Gómez, M.S.; Gerba, C.P.; Bright, K.R. Survival of Respiratory Viruses on Fresh Produce. Food Environ. Virol. 2013, 5, 150–156. [Google Scholar] [CrossRef]
- Conway, M. Identification of coronavirus sequences in carp cDNA from Wuhan, China. J. Med. Virol. 2020, 92, 1629–1633. [Google Scholar] [CrossRef]
- Liu, R.; Stanway, D. Traces of Coronavirus Found in Frozen Chicken Wings, Shrimp Packaging in China, Global News. Available online: https://globalnews.ca/news/7271588/coronavirus-traces-frozen-chicken-shrimp-packaging/ (accessed on 15 July 2021).
- Yusha, Z. China’s CDC Experts Investigate Xinfadi Market Three Times, Announce Groundbreaking Virus Tracing Discovery, Global Times. Available online: https://www.globaltimes.cn/content/1192146.shtml (accessed on 15 July 2021).
- Liu, P.; Yang, M.; Zhao, X.; Guo, Y.; Wang, L.; Zhang, J.; Lei, W.; Han, W.; Jiang, F.; Liu, W.J.; et al. Cold-chain transportation in the frozen food industry may have caused a recurrence of COVID-19 cases in destination: Successful isolation of SARS-CoV-2 virus from the imported frozen cod package surface. Biosaf. Health. 2020, 2, 199–201. [Google Scholar] [CrossRef]
- Pang, X.; Ren, L.; Wu, S.; Ma, W.; Yang, J.; Di, L.; Li, J.; Xiao, Y.; Kang, L.; Du, S.; et al. Cold-chain food contamination as the possible origin of Covid-19 resurgence in Beijing. Natl. Sci. Rev. 2020, 7, 1861–1864. [Google Scholar] [CrossRef]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.; Wang, P.; Duan, F.; Yaron, T.M.; Zhang, T.; Uhl, S.; Bram, Y.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar] [CrossRef]
- Potasman, I.; Paz, A.; Odeh, M. Infectious Outbreaks Associated with Bivalve Shellfish Consumption: A Worldwide Perspective. Clin. Infect. Dis. 2002, 35, 921–928. [Google Scholar] [CrossRef] [Green Version]
- French Agency for Food, Environmental and Occupational Health & Safety (ANSES). Supplemented Opinion of 9 March 2020 of the French Agency for Food, Environmental and Occupational Health & Safety. 2020. Available online: https://www.anses.fr/en/system/files/SABA2020SA0037EN-1.pdf (accessed on 15 July 2021).
- Smith, J.L. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 2003, 66, 1292–1303. [Google Scholar] [CrossRef]
- Hellenic Food Authority (EFET). Guide for the Restart of Foodservice. Available online: http://efet.gr/files/pdf/other/odigos-epan-estiasis.pdf (accessed on 15 July 2021). In Greek.
- Okumus, B. Norovirus and Coronavirus Risks in Food Service Settings: A Systematic Review for Future Research. J. Culin. Sci. Technol. 2021. [Google Scholar] [CrossRef]
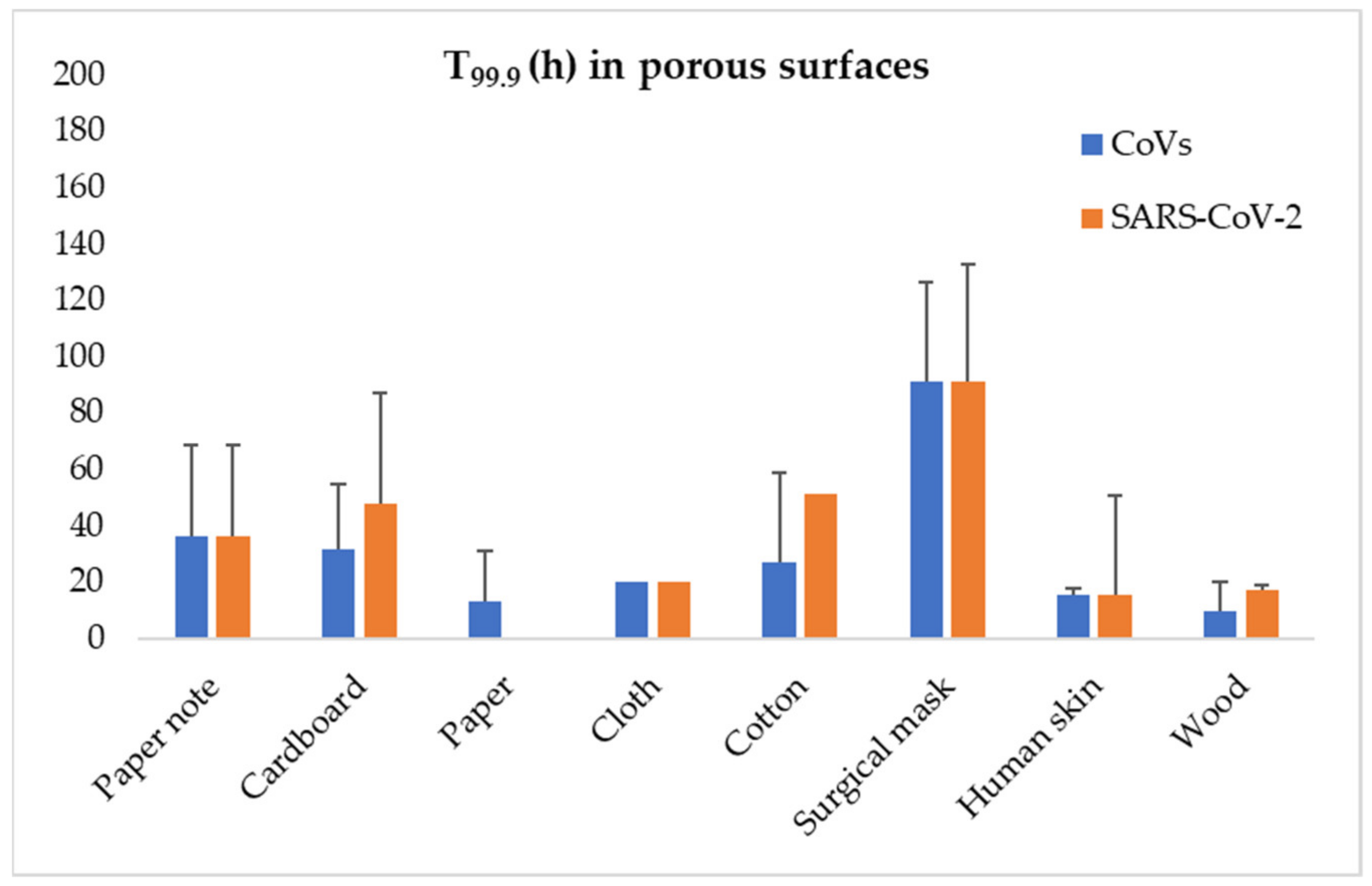
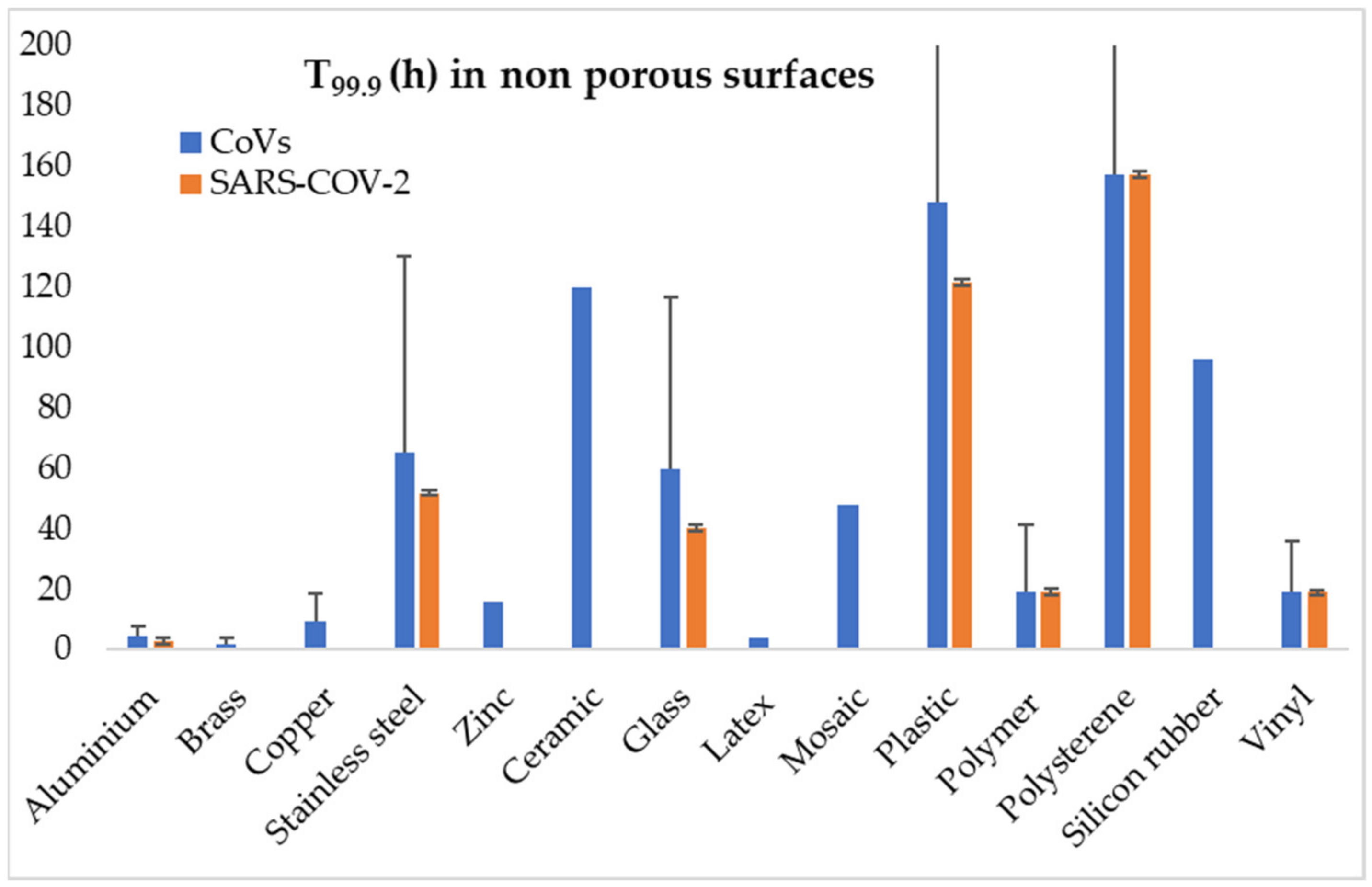
| CoV | Genus | Animal Pool | Intermediate Host | Primary Receptor | Common Transmission | Faecal Shedding | Foodborne Transmission |
|---|---|---|---|---|---|---|---|
| HCoV-229E | alpha-CoV | Bats | Camelids? | Human Aminopeptidase N (CD13) | Respiratory droplets, fomites | ||
| HCoV-OC43 | beta-CoV, lineage A | Rodents | Bovines | 9-O-Acetylated sialic acid | Respiratory droplets, fomites | ||
| SARS-CoV | beta-CoV, lineage B | Palm Civets, Bats | Palm civets | Angiotensin-converting enzyme 2 (ACE2) | Respiratory droplets, fomites, faecal—oral | Yes | Yes |
| HCoV-NL63 | alpha-CoV | Palm Civets, Bats | ? | Angiotensin-converting enzyme 2 (ACE2) | Respiratory droplets, fomites | ||
| HCoV-HKU1 | beta-CoV, lineage A | Rodents | 9-O-Acetylated sialic acid | Respiratory droplets, fomites | |||
| MERS-CoV | beta-CoV, lineage C | Bats, Camels | Dromedary camels | Dipeptidyl peptidase-4 (DPP4) | Respiratory droplets, fomites | Yes | Yes |
| SARS-CoV-2 | beta-CoV, lineage B | Bats | Pangolins? | Angiotensin-converting enzyme 2 (ACE2) | Respiratory droplets, fomites, faecal—oral | Yes | ? |
| Reference | CoV | Water Characteristics | Reduction | |||||
|---|---|---|---|---|---|---|---|---|
| Type | T | pH | Turbidity | 99.9% | 99% | 90% | ||
| Casanova et al., 2009 [88] | TGEV | reagent-grade water | 25 °C | 6 | 0.1 NTU | 33 d | 22 d | 11 d |
| MHV | 26 d | 17 d | 9 d | |||||
| TGEV | 4 °C | 330 d | 220 d | 110 d | ||||
| MHV | >365 d | |||||||
| TGEV | lake water | 25 °C | 7.5 | 1.73 NTU | 13 d | |||
| MHV | 10 d | |||||||
| TGEV | 4 °C | 14 d | ||||||
| MHV | >14 d | |||||||
| Gundy et al., 2019 [87] | HCoV 229E (ATCC-740) | Tap water filtered | 23 °C | 7.8 | 10.1 d | 6.76 d | ||
| FIPV (ATCC-990) | 10.1 d | 6.76 d | ||||||
| HCoV 229E (ATCC-740) | 4 °C | 588 d | 392 d | |||||
| FIPV (ATCC-990) | 130 d | 87 d | ||||||
| HCoV 229E (ATCC-740) | Tap water unfiltered | 23 °C | 12.1 d | 8.09 d | ||||
| FIPV (ATCC-990) | 12.5 d | 8.32 d | ||||||
| Bivins et al., 2020 [89] | SARS-CoV-2 (nCoV-WA1-2020) 10 5 and 10 3 TCID50 *mL−1 | wastewater | 20 °C | 7.98 | 1.6–2.1 d | |||
| tap water | 2.0 d | |||||||
| De Oliveira et al., 2021 [90] | SARS-CoV-2 (SARS.CoV2/SP02. 2020.HIAE.Br) 2 × 10 4 PFU/mL | River water | 24 °C | 5.5 | 10 NTU | 6.4 d | 1.9 d | |
| 4 °C | 18.7 d | 7.7 d | ||||||
| Filtered river water | 24 °C | 1 NTU | 8.5 d | 3.3 d | ||||
| Sala-Comorera et al., 2021 [86] | SARS-CoV-2 3.16 × 10 4 TCID50 *mL−1 | River water | 20 °C | 11.3 d * | 7.5 d * | 3.8 d * | ||
| 4 °C | 11.2 d * | 7.5 d * | 3.7 d * | |||||
| Seawater | 20 °C | 10.5 d * | 7.0 d * | 3.5 d * | ||||
| 4 °C | 5.2 d * | 3.5 d * | 1.7 d * | |||||
| Reference | CoV | Water Characteristics | Reduction | Survival | |||||
|---|---|---|---|---|---|---|---|---|---|
| Type | T | pH | Turbidity | 99.9% | 99% | 90% | |||
| Wang et al., 2005 [77] | SARS-COV-1 | Sewage | 20 °C | 2 d | |||||
| 4 °C | 14 d | ||||||||
| Casanova et al., 2009 [88] | TGEV | Pasteurised sewage | 25 °C | 7.6 | 17.6 NTU | 14 d | 9 d | 4 d | |
| MHV | 10 d | 7 d | 3 d | ||||||
| TGEV | 4 °C | 73 d | 49 d | 24 d | |||||
| MHV | 105 d | 70 d | 35 d | ||||||
| Ye et al., 2016 [3] | MHV A59 | Wastewater | 25 °C | 13 h | |||||
| 10 °C | 36 h | ||||||||
| Pasteurized wastewater | 25 °C | 19 h | |||||||
| Gundy et al., 2019 [87] | HCoV 229E (ATCC-740) | Primary effluent unfiltered | 10 °C | 3.54 d | 2.36 d | ||||
| FIPV (ATCC-990) | 2.56 d | 1.71 d | |||||||
| HCoV 229E (ATCC-740) | Primary effluent filtered | 2.35 d | 1.57 d | ||||||
| FIPV (ATCC-990) | 2.4 d | 1.6 d | |||||||
| HCoV 229E (ATCC-740) | Secondary effluent | 2.77 d | 1.85 d | ||||||
| FIPV (ATCC-990) | 2.42 d | 1.62 d | |||||||
| De Oliveira et al., 2021 [90] | SARS-CoV-2 (SARS.CoV2/SP02. 2020.HIAE.Br) 2 × 10 4 PFU/mL | Wastewater | 24 °C | 7.5 | 274 NTU | 4.0 d | 1.2 d | ||
| 4 °C | 17.5 d | 5.5 d | |||||||
| Filtered wastewater | 24 °C | 6 NTU | 4.5 d | 1.5 d | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Economou, V.; Sakkas, H.; Bezirtzoglou, E.; Papa, A.; Soultos, N. SARS–CoV–2 and Food—How Confident Are We about Them? Hygiene 2021, 1, 80-98. https://0-doi-org.brum.beds.ac.uk/10.3390/hygiene1030008
Economou V, Sakkas H, Bezirtzoglou E, Papa A, Soultos N. SARS–CoV–2 and Food—How Confident Are We about Them? Hygiene. 2021; 1(3):80-98. https://0-doi-org.brum.beds.ac.uk/10.3390/hygiene1030008
Chicago/Turabian StyleEconomou, Vangelis, Hercules Sakkas, Eugenia Bezirtzoglou, Anna Papa, and Nikolaos Soultos. 2021. "SARS–CoV–2 and Food—How Confident Are We about Them?" Hygiene 1, no. 3: 80-98. https://0-doi-org.brum.beds.ac.uk/10.3390/hygiene1030008








