Oxygen Is Instrumental for Biological Signaling: An Overview
Abstract
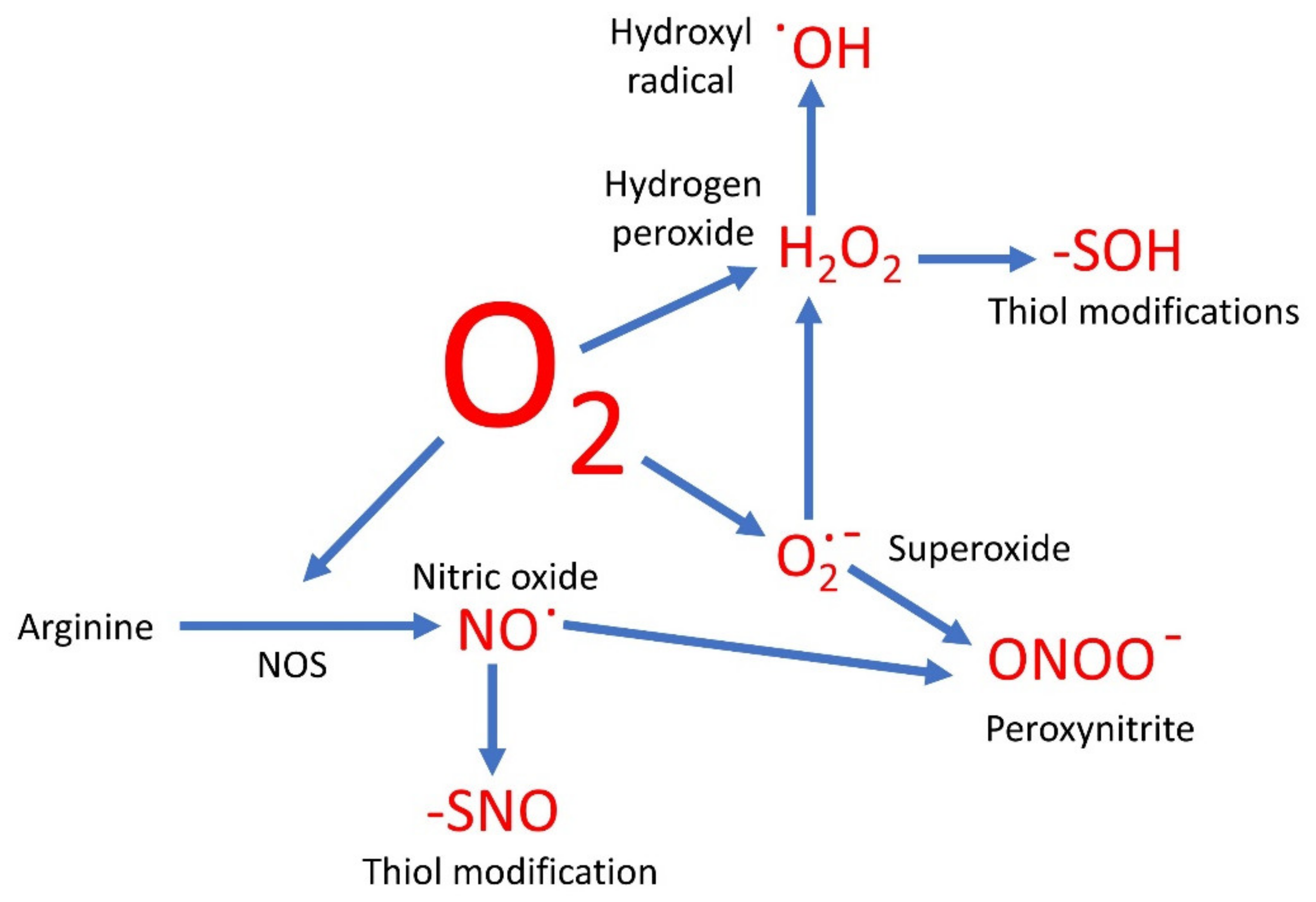
:1. Introduction
2. Signaling by ROS
2.1. Superoxide and Its Role
2.2. Hydrogen Peroxide as a Signal
2.3. Hydroxyl Radicals Can Be Signals Too
3. Signaling by RNS
3.1. Nitric Oxide and Working with Other Oxygen-Based Molecules
3.2. Peroxynitrite, as a Signal
4. The Signaling of Carbon Monoxide
5. Discussion and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lindermayr, C. Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef]
- Beckmann, R.; Flohé, L. The pathogenic role of superoxide radicals in inflammation: Efficacy of exogenous superoxide dismutase. Bull. Eur. Physiopathol. Respir. 1981, 17, 275–286. [Google Scholar]
- Hohn, D.C.; Lehrer, R.I. NADPH oxidase deficiency in X-linked chronic granulomatous disease. J. Clin. Investig. 1975, 55, 707–713. [Google Scholar] [CrossRef] [Green Version]
- Arnold, D.E.; Heimall, J.R. A review of Chronic Granulomatous Disease. Adv. Ther. 2017, 34, 2543–2557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, R.M.; Ferrige, A.G.; Moncada, S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987, 327, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Anson, M.L.; Mirsky, A.E. On the combination of nitric oxide with haemoglobin. J. Physiol. 1925, 60, 100–102. [Google Scholar] [CrossRef] [Green Version]
- Brandes, R.P.; Weissmann, N.; Schröder, K. Nox family NADPH oxidases: Molecular mechanisms of activation. Free Radic. Biol. Med. 2014, 76, 208–226. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef] [PubMed]
- El-Bahr, S.M. Biochemistry of free radicals and oxidative stress. Sci. Int. 2013, 1, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Speckmann, B.; Steinbrenner, H.; Grune, T.; Klotz, L.O. Peroxynitrite: From interception to signaling. Arch. Biochem. Biophys. 2016, 595, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.; Sethi, J.; Choi, A.M. Carbon monoxide-dependent signaling. Crit. Care Med. 2002, 30 Suppl. 1, S12–S17. [Google Scholar] [CrossRef] [PubMed]
- Olas, B. Hydrogen sulfide in signaling pathways. Clin. Chim. Acta. 2015, 439, 212–218. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, M.; Sun, X. Molecular hydrogen is involved in phytohormone signaling and stress responses in plants. PLoS ONE 2013, 8, e71038. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Whiteman, M. Hydrogen sulfide and cell signaling: Team player or referee? Plant Physiol. Biochem. 2014, 78, 37–42. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Cañas, A.; Palma, J.M. Nitric oxide and hydrogen sulfide in plants: Which comes first? J. Exp. Bot. 2019, 70, 4391–4404. [Google Scholar] [CrossRef]
- Monteiro, H.P.; Rodrigues, E.G.; Amorim Reis, A.K.C.; Longo, L.S., Jr.; Ogata, F.T.; Moretti, A.I.S.; da Costa, P.E.; Teodoro, A.C.S.; Toledo, M.S.; Stern, A. Nitric oxide and interactions with reactive oxygen species in the development of melanoma, breast, and colon cancer: A redox signaling perspective. Nitric Oxide 2019, 89, 1–13. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Brand, M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Kettle, A.J.; Hampton, M.B. Reactive oxygen species and neutrophil function. Annu. Rev. Biochem. 2016, 85, 765–792. [Google Scholar] [CrossRef]
- Schröder, K. NADPH oxidases: Current aspects and tools. Redox Biol. 2020. [Google Scholar] [CrossRef]
- Qu, Y.; Yan, M.; Zhang, Q. Functional regulation of plant NADPH oxidase and its role in signaling. Plant Signal. Behav. 2017, 12, e1356970. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.M.; Muhlemann, J.K.; Gayomba, S.R.; Muday, G.K. RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem. Res. Toxicol. 2019, 32, 370–396. [Google Scholar] [CrossRef]
- Lambeth, J.D. Nox/Duox family of nicotinamide adenine dinucleotide (phosphate) oxidases. Curr. Opin. Hematol. 2002, 9, 11–17. [Google Scholar] [CrossRef]
- Kámán-Tóth, E.; Dankó, T.; Gullner, G.; Bozsó, Z.; Palkovics, L.; Pogány, M. Contribution of cell wall peroxidase- and NADPH oxidase-derived reactive oxygen species to Alternaria brassicicola-induced oxidative burst in Arabidopsis. Mol. Plant Pathol. 2019, 20, 485–499. [Google Scholar] [CrossRef] [Green Version]
- Colton, C.; Yao, J.; Grossman, Y.; Gilbert, D. The effect of xanthine/xanthine oxidase generated reactive oxygen species on synaptic transmission. Free Radic. Res. Commun. 1991, 14, 385–393. [Google Scholar] [CrossRef]
- Millar, T.M.; Stevens, C.R.; Benjamin, N.; Eisenthal, R.; Harrison, R.; Blake, D.R. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett. 1998, 427, 225–228. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Arasimowicz, M.; Floryszak-Wieczorek, J. Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Sci. 2007, 172, 876–887. [Google Scholar] [CrossRef]
- Liochev, S.I. Reactive oxygen species and the free radical theory of aging. Free Radic. Biol. Med. 2013, 60, 1–4. [Google Scholar] [CrossRef]
- Lapointe, J.; Hekimi, S. When a theory of aging ages badly. Cell. Mol. Life Sci. 2010, 67, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harman, D. The biologic clock: The mitochondria? J. Am. Geriatr. Soc. 1972, 20, 145–147. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Dec, K.; Kałduńska, J.; Kawczuga, D.; Kochman, J.; Janda, K. Reactive oxygen species-sources, functions, oxidative damage. Pol. Merkur. Lek. Organ Pol. Tow. Lek. 2020, 48, 124–127. [Google Scholar]
- Ma, J.; Zhou, H.; Yan, S.; Song, W. Kinetics studies and mechanistic considerations on the reactions of superoxide radical ions with dissolved organic matter. Water Res. 2019, 149, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Wegerich, F.; Turano, P.; Allegrozzi, M.; Möhwald, H.; Lisdat, F. Cytochrome C mutants for superoxide biosensors. Anal. Chem. 2009, 81, 2976–2984. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef] [Green Version]
- Ewald, C.Y. Redox signaling of NADPH oxidases regulates oxidative stress responses, immunity and aging. Antioxidants 2018, 7, 130. [Google Scholar] [CrossRef] [Green Version]
- Buetler, T.M.; Krauskopf, A.; Ruegg, U.T. Role of superoxide as a signaling molecule. News Physiol. Sci. 2004, 19, 120–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandel, N.S.; McClintock, D.S.; Feliciano, C.E.; Wood, T.M.; Melendez, J.A.; Rodriguez, A.M.; Schumacker, P.T. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1 alpha during hypoxia: A mechanism of O2 sensing. J. Biol. Chem. 2000, 275, 25130–25138. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Lai, U.H.; Zhu, L.; Singh, A.; Ahmed, M.; Forsyth, N.R. Reactive oxygen species formation in the brain at different oxygen levels: The role of hypoxia inducible factors. Front. Cell Dev. Biol. 2018, 6, 132. [Google Scholar] [CrossRef] [Green Version]
- Wood, P.M. The potential diagram for oxygen at pH 7. Biochem. J. 1988, 253, 287–289. [Google Scholar] [CrossRef]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Hancock, J.T. Considerations of the importance of redox state for reactive nitrogen species action. J. Exp. Bot. 2019, 70, 4323–4331. [Google Scholar] [CrossRef] [PubMed]
- Wojtovich, A.P.; Berry, B.J.; Galkin, A. Redox signaling through compartmentalization of reactive oxygen species: Implications for health and disease. Antioxid. Redox Signal. 2019, 31, 591–593. [Google Scholar] [CrossRef] [PubMed]
- Aratani, Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch. Biochem. Biophys. 2018, 640, 47–52. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, C.; Calderon, P.B. Catalase, a remarkable enzyme: Targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 2017, 398, 1095–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, S.G. Overview on peroxiredoxin. Mol. Cells 2016, 39, 1. [Google Scholar]
- Brigelius-Flohé, R.; Flohé, L. Regulatory phenomena in the glutathione peroxidase superfamily. Antioxid. Redox Signal. 2020, 33, 498–516. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winterbourn, C.C. Hydrogen peroxide reactivity and specificity in thiol-based cell signalling. Biochem. Soc. Trans. 2020, 48, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Baty, J.W.; Hampton, M.B.; Winterbourn, C.C. Proteomic detection of hydrogen peroxide-sensitive thiol proteins in Jurkat cells. Biochem. J. 2005, 389, 785–795. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, C.L.; Davies, M.J. Detection, identification, and quantification of oxidative protein modifications. J. Biol. Chem. 2019, 294, 19683–19708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmeen, A.; Andersen, J.N.; Myers, M.P.; Meng, T.C.; Hinks, J.A.; Tonks, N.K.; Barford, D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 2003, 423, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, S.; Park, J.B.; Lee, S.D.; Kim, J.H.; Ha, S.H.; Hasumi, K.; Endo, A.; Suh, P.G.; Ryu, S.H. Hydrogen peroxide induces association between glyceraldehyde 3-phosphate dehydrogenase and phospholipase D2 to facilitate phospholipase D2 activation in PC12 cells. J. Neurochem. 2003, 85, 1228–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, T.; Knuesting, J.; Berndt, C.; Morgan, B.; Scheibe, R. Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol. Chem. 2015, 396, 523–537. [Google Scholar] [CrossRef]
- Desikan, R.; A.-H.-Mackerness, S.; Hancock, J.T.; Neill, S.J. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001, 127, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.C.; Arany, P.R.; Huang, Y.Y.; Tomkinson, E.M.; Sharma, S.K.; Kharkwal, G.B.; Saleem, T.; Mooney, D.; Yull, F.E.; Blackwell, T.S.; et al. Low-level laser therapy activates NF-kB via generation of reactive oxygen species in mouse embryonic fibroblasts. PLoS ONE 2011, 6, e22453. [Google Scholar]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [Green Version]
- Kohlgrüber, S.; Upadhye, A.; Dyballa-Rukes, N.; McNamara, C.A.; Altschmied, J. Regulation of transcription factors by reactive oxygen species and nitric oxide in vascular physiology and pathology. Antioxid. Redox Signal. 2017, 26, 679–699. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Oka, S.; Ale-Agha, N.; Eaton, P. Hydrogen peroxide sensing and signaling by protein kinases in the cardiovascular system. Antioxid. Redox Signal. 2013, 18, 1042–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roscoe, J.M.; Sevier, C.S. Pathways for sensing and responding to hydrogen peroxide at the endoplasmic reticulum. Cells 2020, 9, 2314. [Google Scholar] [CrossRef] [PubMed]
- Veal, E.A.; Day, A.M.; Morgan, B.A. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007, 26, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Bae, S.; Choi, K.H.; An, S. Hydrogen peroxide controls Akt activity via ubiquitination/degradation pathways. Oncol. Rep. 2011, 26, 1561–1566. [Google Scholar]
- Gaschler, M.M.; Stockwell, B.R. Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 2017, 482, 419–425. [Google Scholar] [CrossRef]
- Watt, B.E.; Proudfoot, A.T.; Vale, J.A. Hydrogen peroxide poisoning. Toxicol. Rev. 2004, 23, 51–57. [Google Scholar] [CrossRef]
- Gechev, T.S.; Hille, J. Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 2005, 168, 17–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Zhang, Y.N.; Jia, Q.Q.; Ji, A.; Shao, S.X.; Zhang, L.; Gong, M.; Yin, Q.; Huang, X.L. MicroRNA-214 protects L6 skeletal myoblasts against hydrogen peroxide-induced apoptosis. Free Radic. Res. 2020, 54, 162–172. [Google Scholar] [CrossRef]
- Sies, H. Role of metabolic H2O2 generation: Redox signaling and oxidative stress. J. Biol. Chem. 2014, 289, 8735–8741. [Google Scholar] [CrossRef] [Green Version]
- Alleman, R.J.; Katunga, L.A.; Nelson, M.A.; Brown, D.A.; Anderson, E.J. The “Goldilocks Zone” from a redox perspective-Adaptive vs. deleterious responses to oxidative stress in striated muscle. Front. Physiol. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Manford, A.G.; Rodríguez-Pérez, F.; Shih, K.Y.; Shi, Z.; Berdan, C.A.; Choe, M.; Titov, D.V.; Nomura, D.K.; Rape, M. A cellular mechanism to detect and alleviate reductive stress. Cell 2020, 183, 46–61. [Google Scholar] [CrossRef]
- Hancock, J.T.; Veal, D. Nitric oxide, other reactive signalling compounds, redox, and reductive stress. J. Exp. Bot. 2021, 72, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Fischbacher, A.; von Sonntag, C.; Schmidt, T.C. Hydroxyl radical yields in the Fenton process under various pH, ligand concentrations and hydrogen peroxide/Fe(II) ratios. Chemosphere 2017, 182, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Kehrer, J.P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Fong, K.L.; McCay, P.B.; Poyer, J.L. Evidence for superoxide-dependent reduction of Fe3+ and its role in enzyme-generated hydroxyl radical formation. Chem. Biol. Interact. 1976, 15, 77–89. [Google Scholar] [CrossRef]
- Halliwell, B. Superoxide-dependent formation of hydroxyl radicals in the presence of iron chelates: Is it a mechanism for hydroxyl radical production in biochemical systems? FEBS Lett. 1978, 92, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Richards, S.L.; Wilkins, K.A.; Swarbreck, S.M.; Anderson, A.A.; Habib, N.; Smith, A.G.; McAinsh, M.; Davies, J.M. The hydroxyl radical in plants: From seed to seed. J. Exp. Bot. 2015, 66, 37–46. [Google Scholar] [CrossRef]
- Pottosin, I.; Zepeda-Jazo, I.; Bose, J.; Shabala, S. An anion conductance, the essential component of the hydroxyl-radical-induced ion current in plant roots. Int. J. Mol. Sci. 2018, 19, 897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demidchik, V.; Cuin, T.A.; Svistunenko, D.; Smith, S.J.; Miller, A.J.; Shabala, S.; Sokolik, A.; Yurin, V. Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: Single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 2010, 123, 1468–1479. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.J.; Lin, K.H.; Hsu, M.J.; Chou, D.S.; Hsiao, G.; Sheu, J.R. Suppression of NF-κB signaling by andrographolide with a novel mechanism in human platelets: Regulatory roles of the p38 MAPK-hydroxyl radical-ERK2 cascade. Biochem. Pharmacol. 2012, 84, 914–924. [Google Scholar] [CrossRef]
- Chen, W.; Ding, S.; Wu, J.; Shi, G.; Zhu, A. In situ detection of hydroxyl radicals in mitochondrial oxidative stress with a nanopipette electrode. Chem. Commun. 2020, 56, 13225–13228. [Google Scholar] [CrossRef]
- Sakai, T.; Imai, J.; Ito, T.; Takagaki, H.; Ui, M.; Hatta, S. The novel antioxidant TA293 reveals the role of cytoplasmic hydroxyl radicals in oxidative stress-induced senescence and inflammation. Biochem. Biophys. Res. Commun. 2017, 482, 1183–1189. [Google Scholar] [CrossRef]
- Xu, G.; Chance, M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007, 107, 3514–3543. [Google Scholar] [CrossRef]
- Tejero, I.; Gonzalez-Lafont, A.; Lluch, J.M.; Eriksson, L.A. Theoretical modeling of hydroxyl-radical-induced lipid peroxidation reactions. J. Phys. Chem. B 2007, 111, 5684–5693. [Google Scholar] [CrossRef]
- Gilbert, B.C.; King, D.M.; Thomas, C.B. The oxidation of some polysaccharides by the hydroxyl radical: An e.s.r. investigation. Carbohydr. Res. 1984, 125, 217–235. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Pospíšil, P. Data on detection of singlet oxygen, hydroxyl radical and organic radical in Arabidopsis thaliana. Data Brief. 2018, 21, 2246–2252. [Google Scholar] [CrossRef]
- Shen, B.; Jensen, R.G.; Bohnert, H.J. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 1997, 115, 527–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipinski, B. Hydroxyl radical and its scavengers in health and disease. Oxid. Med. Cell Longev. 2011, 2011, 809696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, S. Molecular hydrogen as a novel antioxidant: Overview of the advantages of hydrogen for medical applications. Methods Enzymol. 2015, 555, 289–317. [Google Scholar]
- Iida, A.; Nosaka, N.; Yumoto, T.; Knaup, E.; Naito, H.; Nishiyama, C.; Yamakawa, Y.; Tsukahara, K.; Terado, M.; Sato, K.; et al. The clinical application of hydrogen as a medical treatment. Acta Med. Okayama 2016, 70, 331–337. [Google Scholar] [PubMed]
- Ohsawa, I.; Ishikawa, M.; Takahashi, K.; Watanabe, M.; Nishimaki, K.; Yamagata, K.; Katsura, K.; Katayama, Y.; Asoh, S.; Ohta, S. Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat. Med. 2007, 13, 688. [Google Scholar] [CrossRef]
- Penders, J.; Kissner, R.; Koppenol, W.H. ONOOH does not react with H2: Potential beneficial effects of H2 as an antioxidant by selective reaction with hydroxyl radicals and peroxynitrite. Free Radic. Biol. Med. 2014, 75, 191–194. [Google Scholar] [CrossRef]
- Hancock, J.T.; Russell, G. Downstream signalling from molecular hydrogen. Plants 2021, 10, 367. [Google Scholar] [CrossRef] [PubMed]
- Bredt, D.S.; Snyder, S.H. Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc. Natl. Acad. Sci. USA 1990, 87, 682–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Tenopoulou, M.; Doulias, P.T. Endothelial nitric oxide synthase-derived nitric oxide in the regulation of metabolism. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Kolbert, Z.; Barroso, J.B.; Brouquisse, R.; Corpas, F.J.; Gupta, K.J.; Lindermayr, C.; Loake, G.J.; Palma, M.; Petřivalský, M.; Wendehenne, D.; et al. A forty year journey: The generation and roles of NO in plants. Nitric Oxide 2019, 93, 53–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshland, D.E. The molecular of the year (Editorial). Science 1992, 258, 1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toda, N.; Ayajiki, K.; Okamura, T. Cerebral blood flow regulation by nitric oxide: Recent advances. Pharmacol. Rev. 2009, 61, 62–97. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.R.; Yue, C.M.; Hao, F.S. Update on roles of nitric oxide in regulating stomatal closure. Plant Signal. Behav. 2019, 14, 1649569. [Google Scholar] [CrossRef] [PubMed]
- Stuehr, D.J.; Vasquez-Vivar, J. Nitric oxide synthases-from genes to function. Nitric Oxide 2017, 63, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Xiao, S.; Li, Q.; Hu, L.; Yu, Z.; Yang, J.; Chang, Q.; Chen, Z.; Hu, G. Soluble guanylate cyclase stimulators and activators: Where are we and where to go? Mini Rev. Med. Chem. 2019, 19, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Mounier, A.; Santolini, J.; Jeandroz, S.; Wendehenne, D. The evolution of nitric oxide signalling diverges between animal and green lineages. J. Exp. Bot. 2019, 70, 4355–4364. [Google Scholar] [CrossRef]
- Feng, J.; Chen, L.; Zuo, J. Protein S-nitrosylation in plants: Current progresses and challenges. J. Integr. Plant Biol. 2019, 61, 1206–1223. [Google Scholar] [CrossRef]
- Gupta, K.J.; Hancock, J.T.; Petrivalsky, M.; Kolbert, Z.; Lindermayr, C.; Durner, J.; Barroso, J.B.; Palma, J.M.; Brouquisse, R.; Wendehenne, D.; et al. Recommendations on terminology and experimental best practice associated with plant nitric oxide research. New Phytol. 2020, 225, 1828–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, A.K.; Gadalla, M.M.; Sen, N.; Kim, S.; Mu, W.; Gazi, S.K.; Barrow, R.K.; Yang, G.; Wang, R.; Snyder, S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009, 2, ra72. [Google Scholar] [CrossRef] [Green Version]
- Aroca, Á.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 2015, 168, 334–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Checconi, P.; Limongi, D.; Baldelli, S.; Ciriolo, M.R.; Nencioni, L.; Palamara, A.T. Role of glutathionylation in infection and inflammation. Nutrients 2019, 11, 1952. [Google Scholar] [CrossRef] [Green Version]
- Kolbert, Z.; Feigl, G.; Bordé, Á.; Molnár, Á.; Erdei, L. Protein tyrosine nitration in plants: Present knowledge, computational prediction and future perspectives. Plant Physiol. Biochem. 2017, 113, 56–63. [Google Scholar] [CrossRef] [Green Version]
- Ventimiglia, L.; Mutus, B. The physiological implications of S-nitrosoglutathione reductase (GSNOR) activity mediating NO signalling in plant root structures. Antioxidants 2020, 9, 1206. [Google Scholar] [CrossRef]
- Rassaf, T.; Preik, M.; Kleinbongard, P.; Lauer, T.; Heiss, C.; Strauer, B.E.; Feelisch, M.; Kelm, M. Evidence for in vivo transport of bioactive nitric oxide in human plasma. J. Clin. Investig. 2002, 109, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, M.; Li, L.; Kostetski, I.; Chu, S.H.; Siau, J.L.; Bhatia, M.; Moore, P.K. Evidence for the formation of a novel nitrosothiol from the gaseous mediators nitric oxide and hydrogen sulphide. Biochem. Biophys. Res. Commun. 2006, 343, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Sun, L.; Liu, M.; Zhou, J. Effect of nitric oxide on reactive oxygen species and antioxidant enzymes in kiwifruit during storage. J. Sci. Food Agric. 2008, 88, 2324–2331. [Google Scholar] [CrossRef]
- Vandelle, E.; Delledonne, M. Peroxynitrite formation and function in plants. Plant Sci. 2011, 181, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, B.; Radi, R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids 2003, 25, 295–311. [Google Scholar] [CrossRef]
- Staszek, P.; Gniazdowska, A. Peroxynitrite induced signaling pathways in plant response to non-proteinogenic amino acids. Planta 2020, 252, 5. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.O.; Schroeder, P.; Sies, H. Peroxynitrite signaling: Receptor tyrosine kinases and activation of stress-responsive pathways. Free Radic. Biol. Med. 2002, 33, 737–743. [Google Scholar] [CrossRef]
- Shacka, J.J.; Sahawneh, M.A.; Gonzalez, J.D.; Ye, Y.Z.; D’alessandro, T.L.; Estevez, A.G. Two distinct signaling pathways regulate peroxynitrite-induced apoptosis in PC12 cells. Cell Death Differ. 2006, 13, 1506–1514. [Google Scholar] [CrossRef] [Green Version]
- Wilks, A. Heme oxygenase: Evolution, structure, and mechanism. Antioxid. Redox Signal. 2002, 4, 603–614. [Google Scholar] [CrossRef]
- Wegiel, B.; Nemeth, Z.; Correa-Costa, M.; Bulmer, A.C.; Otterbein, L.E. Heme oxygenase-1: A metabolic nike. Antioxid. Redox Signal. 2014, 20, 1709–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldbaum, L.R.; Ramirez, R.G.; Absalon, K.B. What is the mechanism of carbon monoxide toxicity? Aviat. Space Environ. Med. 1975, 46, 1289–1291. [Google Scholar]
- Peers, C.; Boyle, J.P.; Scragg, J.L.; Dallas, M.L.; Al-Owais, M.M.; Hettiarachichi, N.T.; Elies, J.; Johnson, E.; Gamper, N.; Steele, D.S. Diverse mechanisms underlying the regulation of ion channels by carbon monoxide. Br. J. Pharmacol. 2015, 172, 1546–1556. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Zhang, W.; Qi, F.; Cui, W.; Xie, Y.; Shen, W. Hydrogen-rich water regulates cucumber adventitious root development in a heme oxygenase-1/carbon monoxide-dependent manner. J. Plant Physiol. 2014, 171, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kourti, M.; Jiang, W.G.; Cai, J. Aspects of carbon monoxide in form of CO-releasing molecules used in cancer treatment: More light on the way. Oxidative Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef]
- Hancock, J.T. Harnessing evolutionary toxins for signaling: Reactive oxygen species, nitric oxide and hydrogen sulfide in plant cell regulation. Front. Plant Sci. 2017, 8, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, H. Reactive oxygen species promote vascular smooth muscle cell proliferation. Circ. Res. 2013, 113, 1040–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Lutsenko, S.V.; Terentiev, A.A. Reactive oxygen and nitrogen species–induced protein modifications: Implication in carcinogenesis and anticancer therapy. Cancer Res. 2018, 78, 6040–6047. [Google Scholar] [CrossRef] [Green Version]
- Turpaev, K.T. Reactive oxygen species and regulation of gene expression. Biochemistry 2002, 67, 281–292. [Google Scholar] [CrossRef]
- Volpe, C.M.O.; Villar-Delfino, P.H.; Dos Anjos, P.M.F.; Nogueira-Machado, J.A. Cellular death, reactive oxygen species (ROS) and diabetic complications. Cell Death Dis. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Collin, F. Chemical basis of reactive oxygen species reactivity and involvement in neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 2407. [Google Scholar] [CrossRef] [Green Version]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res. 2018, 122, 877–902. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Nitro-oxidative stress vs. oxidative or nitrosative stress in higher plants. New Phytol. 2013, 199, 633–635. [Google Scholar] [CrossRef] [PubMed]
- Grisan, F.; Iannucci, L.F.; Surdo, N.C.; Gerbino, A.; Zanin, S.; Di Benedetto, G.; Pozzan, T.; Lefkimmiatis, K. PKA compartmentalization links cAMP signaling and autophagy. Cell Death Differ. 2021, 1–14. [Google Scholar] [CrossRef]
- Petersen, O.H. Calcium signal compartmentalization. Biol. Res. 2002, 35, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hillion, M.; Antelmann, H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015, 396, 415–444. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hancock, J.T. Oxygen Is Instrumental for Biological Signaling: An Overview. Oxygen 2021, 1, 3-15. https://0-doi-org.brum.beds.ac.uk/10.3390/oxygen1010002
Hancock JT. Oxygen Is Instrumental for Biological Signaling: An Overview. Oxygen. 2021; 1(1):3-15. https://0-doi-org.brum.beds.ac.uk/10.3390/oxygen1010002
Chicago/Turabian StyleHancock, John T. 2021. "Oxygen Is Instrumental for Biological Signaling: An Overview" Oxygen 1, no. 1: 3-15. https://0-doi-org.brum.beds.ac.uk/10.3390/oxygen1010002





