TOR Signaling Pathway
Share This Topical Collection
Editors
 Prof. Kazuhiro Shiozaki
Prof. Kazuhiro Shiozaki
 Prof. Kazuhiro Shiozaki
Prof. Kazuhiro Shiozaki
E-Mail
Website
Collection Editor
1. Graduate School of Biological Sciences, Nara Institute of Science and Technology, Ikoma 630-0192, Nara, Japan
2. Department of Microbiology and Molecular Genetics, University of California, Davis, CA 95616, USA
Interests: cellular signal transduction pathways in yeast and humans
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Among the numerous protein kinases that play key roles in signal transduction pathways of eukaryotic cells, Target of Rapamycin (TOR) stands out because of its unique characteristics. TOR is a serine/threonine-specific protein kinase, but it is structurally related to lipid kinases, such as PI3K. It forms at least two distinct high-molecular weight Complexes, known as TOR Complex 1 (TORC1) and TOR Complex 2 (TORC2), with multiple regulatory subunits that determine signal inputs, substrate specificities, and intracellular localization. Rapamycin and other inhibitors of TOR affect diverse aspects of cellular physiology, such as growth, proliferation, as well as catabolic and anabolic processes, suggesting TOR functions at pivotal nodes of cellular signaling networks.
We invite contributions from researchers who have been exploring distinct aspects of this unique protein kinase through studies in diverse model organisms. Both original research articles and reviews are welcome. Together, these studies will contribute to an integrated view of the emerging TOR network, implicated in cancers, metabolic diseases and aging in humans.
Prof. Kazuhiro Shiozaki
Prof. Dr. Ted Powers
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- Target of Rapamycin
- TOR
- mTOR
- rapamycin
Related Special Issue
Published Papers (10 papers)
Open AccessFeature PaperEditor’s ChoiceReview
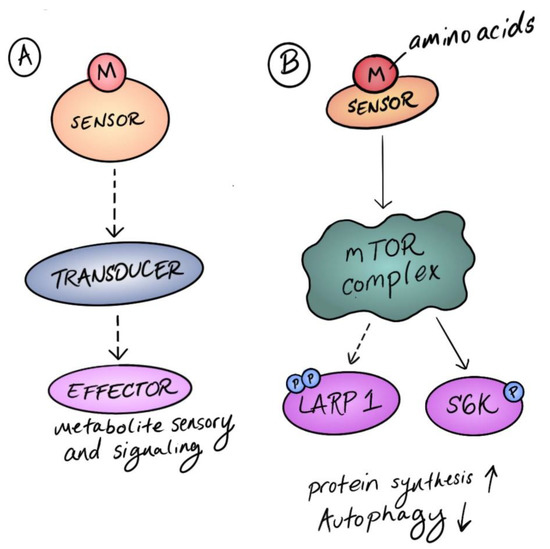
Amino Acid Signaling for TOR in Eukaryotes: Sensors, Transducers, and a Sustainable Agricultural fuTORe
by
Nanticha Lutt and Jacob O. Brunkard
Cited by 11 | Viewed by 3266
Abstract
Eukaryotic cells monitor and regulate metabolism through the atypical protein kinase target of rapamycin (TOR) regulatory hub. TOR is activated by amino acids in animals and fungi through molecular signaling pathways that have been extensively defined in the past ten years. Very recently,
[...] Read more.
Eukaryotic cells monitor and regulate metabolism through the atypical protein kinase target of rapamycin (TOR) regulatory hub. TOR is activated by amino acids in animals and fungi through molecular signaling pathways that have been extensively defined in the past ten years. Very recently, several studies revealed that TOR is also acutely responsive to amino acid metabolism in plants, but the mechanisms of amino acid sensing are not yet established. In this review, we summarize these discoveries, emphasizing the diversity of amino acid sensors in human cells and highlighting pathways that are indirectly sensitive to amino acids, i.e., how TOR monitors changes in amino acid availability without a
bona fide amino acid sensor. We then discuss the relevance of these model discoveries to plant biology. As plants can synthesize all proteinogenic amino acids from inorganic precursors, we focus on the possibility that TOR senses both organic metabolites and inorganic nutrients. We conclude that an evolutionary perspective on nutrient sensing by TOR benefits both agricultural and biomedical science, contributing to ongoing efforts to generate crops for a sustainable agricultural future.
Full article
►▼
Show Figures
Open AccessArticle
RalA, PLD and mTORC1 Are Required for Kinase-Independent Pathways in DGKβ-Induced Neurite Outgrowth
by
Takuya Kano, Ryosuke Tsumagari, Akio Nakashima, Ushio Kikkawa, Shuji Ueda, Minoru Yamanoue, Nobuyuki Takei and Yasuhito Shirai
Viewed by 1507
Abstract
Diacylglycerol kinase β (DGKβ) is an enzyme that converts diacylglycerol to phosphatidic acid and is mainly expressed in the cerebral cortex, hippocampus and striatum. We previously reported that DGKβ induces neurite outgrowth and spinogenesis, contributing to higher brain functions, including emotion and memory.
[...] Read more.
Diacylglycerol kinase β (DGKβ) is an enzyme that converts diacylglycerol to phosphatidic acid and is mainly expressed in the cerebral cortex, hippocampus and striatum. We previously reported that DGKβ induces neurite outgrowth and spinogenesis, contributing to higher brain functions, including emotion and memory. To elucidate the mechanisms involved in neuronal development by DGKβ, we investigated the importance of DGKβ activity in the induction of neurite outgrowth using human neuroblastoma SH-SY5Y cells. Interestingly, both wild-type DGKβ and the kinase-negative (KN) mutant partially induced neurite outgrowth, and these functions shared a common pathway via the activation of mammalian target of rapamycin complex 1 (mTORC1). In addition, we found that DGKβ interacted with the small GTPase RalA and that siRNA against RalA and phospholipase D (PLD) inhibitor treatments abolished DGKβKN-induced neurite outgrowth. These results indicate that binding of RalA and activation of PLD and mTORC1 are involved in DGKβKN-induced neurite outgrowth. Taken together with our previous reports, mTORC1 is a key molecule in both kinase-dependent and kinase-independent pathways of DGKβ-mediated neurite outgrowth, which is important for higher brain functions.
Full article
►▼
Show Figures
Open AccessFeature PaperReview
Pib2 as an Emerging Master Regulator of Yeast TORC1
by
Riko Hatakeyama
Cited by 7 | Viewed by 2808
Abstract
Cell growth is dynamically regulated in response to external cues such as nutrient availability, growth factor signals, and stresses. Central to this adaptation process is the Target of Rapamycin Complex 1 (TORC1), an evolutionarily conserved kinase complex that fine-tunes an enormous number of
[...] Read more.
Cell growth is dynamically regulated in response to external cues such as nutrient availability, growth factor signals, and stresses. Central to this adaptation process is the Target of Rapamycin Complex 1 (TORC1), an evolutionarily conserved kinase complex that fine-tunes an enormous number of cellular events. How upstream signals are sensed and transmitted to TORC1 has been intensively studied in major model organisms including the budding yeast
Saccharomyces cerevisiae. This field recently saw a breakthrough: the identification of yeast phosphatidylInositol(3)-phosphate binding protein 2 (Pib2) protein as a critical regulator of TORC1. Although the study of Pib2 is still in its early days, multiple groups have provided important mechanistic insights on how Pib2 relays nutrient signals to TORC1. There remain, on the other hand, significant gaps in our knowledge and mysteries that warrant further investigations. This is the first dedicated review on Pib2 that summarizes major findings and outstanding questions around this emerging key player in cell growth regulation.
Full article
►▼
Show Figures
Open AccessReview
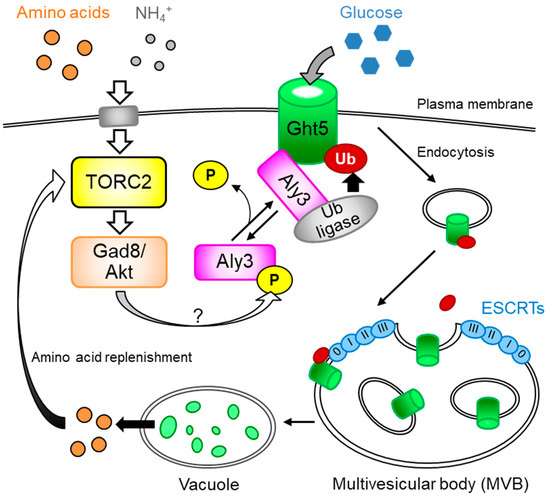
Fission Yeast TORC2 Signaling Pathway Ensures Cell Proliferation under Glucose-Limited, Nitrogen-Replete Conditions
by
Yusuke Toyoda and Shigeaki Saitoh
Cited by 6 | Viewed by 2737
Abstract
Target of rapamycin (TOR) kinases form two distinct complexes, TORC1 and TORC2, which are evolutionarily conserved among eukaryotes. These complexes control intracellular biochemical processes in response to changes in extracellular nutrient conditions. Previous studies using the fission yeast,
Schizosaccharomyces pombe, showed that
[...] Read more.
Target of rapamycin (TOR) kinases form two distinct complexes, TORC1 and TORC2, which are evolutionarily conserved among eukaryotes. These complexes control intracellular biochemical processes in response to changes in extracellular nutrient conditions. Previous studies using the fission yeast,
Schizosaccharomyces pombe, showed that the TORC2 signaling pathway, which is essential for cell proliferation under glucose-limited conditions, ensures cell-surface localization of a high-affinity hexose transporter, Ght5, by downregulating its endocytosis. The TORC2 signaling pathway retains Ght5 on the cell surface, depending on the presence of nitrogen sources in medium. Ght5 is transported to vacuoles upon nitrogen starvation. In this review, we discuss the molecular mechanisms underlying this regulation to cope with nutritional stress, a response which may be conserved from yeasts to mammals.
Full article
►▼
Show Figures
Open AccessArticle
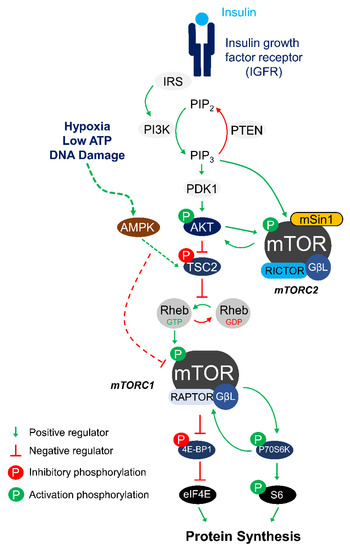
Mechanistic Target of Rapamycin Complex 1 Signaling Links Hypoxia to Increased IGFBP-1 Phosphorylation in Primary Human Decidualized Endometrial Stromal Cells
by
Pinki Nandi, Chloe E. Jang, Kyle Biggar, Chidambra D. Halari, Thomas Jansson and Madhulika B. Gupta
Cited by 3 | Viewed by 2407
Abstract
Insulin-like growth factor-1 (IGF-1) bioavailability in pregnancy is governed by IGF binding protein (IGFBP-1) and its phosphorylation, which enhances the affinity of IGFBP-1 for the growth factor. The decidua is the predominant source of maternal IGFBP-1; however, the mechanisms regulating decidual IGFBP-1 secretion/phosphorylation
[...] Read more.
Insulin-like growth factor-1 (IGF-1) bioavailability in pregnancy is governed by IGF binding protein (IGFBP-1) and its phosphorylation, which enhances the affinity of IGFBP-1 for the growth factor. The decidua is the predominant source of maternal IGFBP-1; however, the mechanisms regulating decidual IGFBP-1 secretion/phosphorylation are poorly understood. Using decidualized primary human endometrial stromal cells (HESCs) from first-trimester placenta, we tested the hypothesis that mTORC1 signaling mechanistically links hypoxia to decidual IGFBP-1 secretion/phosphorylation. Hypoxia inhibited mechanistic target of rapamycin (mTORC1) (p-P70-S6K/Thr389, −47%,
p = 0.038; p-4E-BP1/Thr70, −55%,
p = 0.012) and increased IGFBP-1 (total, +35%,
p = 0.005; phosphorylated, Ser101/+82%,
p = 0.018; Ser119/+88%,
p = 0.039; Ser 169/+157%,
p = 0.019). Targeted parallel reaction monitoring-mass spectrometry (PRM-MS) additionally demonstrated markedly increased dual IGFBP-1 phosphorylation (pSer98+Ser101; pSer169+Ser174) in hypoxia. IGFBP-1 hyperphosphorylation inhibited IGF-1 receptor autophosphorylation/ Tyr1135 (−29%,
p = 0.002). Furthermore, silencing of tuberous sclerosis complex 2 (TSC2) activated mTORC1 (p-P70-S6K/Thr389, +68%,
p = 0.038; p-4E-BP1/Thr70, +30%,
p = 0.002) and reduced total/site-specific IGFBP-1 phosphorylation. Importantly, TSC2 siRNA prevented inhibition of mTORC1 and the increase in secretion/site-specific IGFBP-1 phosphorylation in hypoxia. PRM-MS indicated concomitant changes in protein kinase autophosphorylation (CK2/Tyr182; PKC/Thr497; PKC/Ser657). Overall, mTORC1 signaling mechanistically links hypoxia to IGFBP-1 secretion/phosphorylation in primary HESC, implicating decidual mTORC1 inhibition as a novel mechanism linking uteroplacental hypoxia to fetal growth restriction.
Full article
►▼
Show Figures
Open AccessReview
mTOR Signaling in Metabolic Stress Adaptation
by
Cheng-Wei Wu and Kenneth B. Storey
Cited by 17 | Viewed by 3880
Abstract
The mechanistic target of rapamycin (mTOR) is a central regulator of cellular homeostasis that integrates environmental and nutrient signals to control cell growth and survival. Over the past two decades, extensive studies of mTOR have implicated the importance of this protein complex in
[...] Read more.
The mechanistic target of rapamycin (mTOR) is a central regulator of cellular homeostasis that integrates environmental and nutrient signals to control cell growth and survival. Over the past two decades, extensive studies of mTOR have implicated the importance of this protein complex in regulating a broad range of metabolic functions, as well as its role in the progression of various human diseases. Recently, mTOR has emerged as a key signaling molecule in regulating animal entry into a hypometabolic state as a survival strategy in response to environmental stress. Here, we review current knowledge of the role that mTOR plays in contributing to natural hypometabolic states such as hibernation, estivation, hypoxia/anoxia tolerance, and dauer diapause. Studies across a diverse range of animal species reveal that mTOR exhibits unique regulatory patterns in an environmental stressor-dependent manner. We discuss how key signaling proteins within the mTOR signaling pathways are regulated in different animal models of stress, and describe how each of these regulations uniquely contribute to promoting animal survival in a hypometabolic state.
Full article
►▼
Show Figures
Open AccessArticle
Rapamycin Improves Spatial Learning Deficits, Vulnerability to Alcohol Addiction and Altered Expression of the GluN2B Subunit of the NMDA Receptor in Adult Rats Exposed to Ethanol during the Neonatal Period
by
Malgorzata Lopatynska-Mazurek, Anna Antolak, Pawel Grochecki, Ewa Gibula-Tarlowska, Anna Bodzon-Kulakowska, Joanna Listos, Ewa Kedzierska, Piotr Suder, Jerzy Silberring and Jolanta H. Kotlinska
Cited by 9 | Viewed by 2393
Abstract
Ethanol exposure during pregnancy alters the mammalian target of rapamycin (mTOR) signaling pathway in the fetal brain. Hence, in adult rats exposed to ethanol during the neonatal period, we investigated the influence of rapamycin, an mTOR Complex 1 (mTORC1) inhibitor, on deficits in
[...] Read more.
Ethanol exposure during pregnancy alters the mammalian target of rapamycin (mTOR) signaling pathway in the fetal brain. Hence, in adult rats exposed to ethanol during the neonatal period, we investigated the influence of rapamycin, an mTOR Complex 1 (mTORC1) inhibitor, on deficits in spatial memory and reversal learning in the Barnes maze task, as well as the ethanol-induced rewarding effects (1.0 or 1.5 g/kg) using the conditioning place preference (CPP) paradigm. Rapamycin (3 and 10 mg/kg) was given before intragastric ethanol (5 g/kg/day) administration at postnatal day (PND)4–9 (an equivalent to the third trimester of human pregnancy). Spatial memory/reversal learning and rewarding ethanol effect were evaluated in adult (PND60–70) rats. Additionally, the impact of rapamycin pre-treatment on the expression of the GluN2B subunit of NMDA receptor in the brain was assessed in adult rats. Our results show that neonatal ethanol exposure induced deficits in spatial memory and reversal learning in adulthood, but the reversal learning outcome may have been due to spatial learning impairments rather than cognitive flexibility impairments. Furthermore, in adulthood the ethanol treated rats were also more sensitive to the rewarding effect of ethanol than the control group. Rapamycin prevented the neonatal effect of ethanol and normalized the GluN2B down-regulation in the hippocampus and the prefrontal cortex, as well as normalized this subunit’s up-regulation in the striatum of adult rats. Our results suggest that rapamycin and related drugs may hold promise as a preventive therapy for fetal alcohol spectrum disorders.
Full article
►▼
Show Figures
Open AccessReview
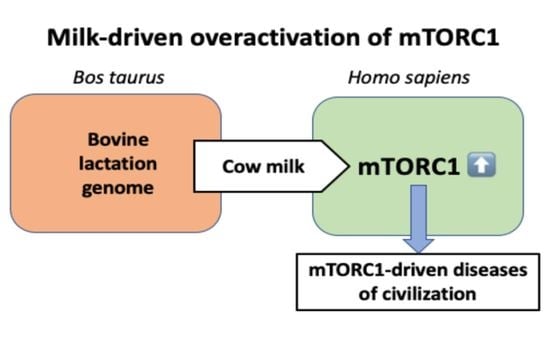
Lifetime Impact of Cow’s Milk on Overactivation of mTORC1: From Fetal to Childhood Overgrowth, Acne, Diabetes, Cancers, and Neurodegeneration
by
Bodo C. Melnik
Cited by 22 | Viewed by 10429
Abstract
The consumption of cow’s milk is a part of the basic nutritional habits of Western industrialized countries. Recent epidemiological studies associate the intake of cow’s milk with an increased risk of diseases, which are associated with overactivated mechanistic target of rapamycin complex 1
[...] Read more.
The consumption of cow’s milk is a part of the basic nutritional habits of Western industrialized countries. Recent epidemiological studies associate the intake of cow’s milk with an increased risk of diseases, which are associated with overactivated mechanistic target of rapamycin complex 1 (mTORC1) signaling. This review presents current epidemiological and translational evidence linking milk consumption to the regulation of mTORC1, the master-switch for eukaryotic cell growth. Epidemiological studies confirm a correlation between cow’s milk consumption and birthweight, body mass index, onset of menarche, linear growth during childhood, acne vulgaris, type 2 diabetes mellitus, prostate cancer, breast cancer, hepatocellular carcinoma, diffuse large B-cell lymphoma, neurodegenerative diseases, and all-cause mortality. Thus, long-term persistent consumption of cow’s milk increases the risk of mTORC1-driven diseases of civilization. Milk is a highly conserved, lactation genome-controlled signaling system that functions as a maternal-neonatal relay for optimized species-specific activation of mTORC1, the nexus for regulation of eukaryotic cell growth, and control of autophagy. A deeper understanding of milk´s impact on mTORC1 signaling is of critical importance for the prevention of common diseases of civilization.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceArticle
Rapamycin Improves Recognition Memory and Normalizes Amino-Acids and Amines Levels in the Hippocampal Dentate Gyrus in Adult Rats Exposed to Ethanol during the Neonatal Period
by
Malgorzata Lopatynska-Mazurek, Anna Pankowska, Ewa Gibula-Tarlowska, Radoslaw Pietura and Jolanta H. Kotlinska
Cited by 13 | Viewed by 2375
Abstract
The mammalian target of rapamycin (mTOR), a serine/ threonine kinase, is implicated in synaptic plasticity by controlling protein synthesis. Research suggests that ethanol exposure during pregnancy alters the mTOR signaling pathway in the fetal hippocampus. Thus, we investigated the influence of pre-treatment with
[...] Read more.
The mammalian target of rapamycin (mTOR), a serine/ threonine kinase, is implicated in synaptic plasticity by controlling protein synthesis. Research suggests that ethanol exposure during pregnancy alters the mTOR signaling pathway in the fetal hippocampus. Thus, we investigated the influence of pre-treatment with rapamycin, an mTORC1 inhibitor, on the development of recognition memory deficits in adult rats that were neonatally exposed to ethanol. In the study, male and female rat pups received ethanol (5 g/kg/day) by intragastric intubation at postanatal day (PND 4-9), an equivalent to the third trimester of human pregnancy. Rapamycin (3 and 10 mg/kg) was given intraperitoneally before every ethanol administration. Short- and long-term recognition memory was assessed in the novel object recognition (NOR) task in adult (PND 59/60) rats. Locomotor activity and anxiety-like behavior were also evaluated to exclude the influence of such behavior on the outcome of the memory task. Moreover, the effects of rapamycin pre-treatment during neonatal ethanol exposure on the content of amino-acids and amines essential for the proper development of cognitive function in the dentate gyrus (DG) of the hippocampus was evaluated using proton magnetic resonance spectroscopy (1H MRS) in male adult (PND 60) rats. Our results show the deleterious effect of ethanol given to neonatal rats on long-term recognition memory in adults. The effect was more pronounced in male rather than female rats. Rapamycin reversed this ethanol-induced memory impairment and normalized the levels of amino acids and amines in the DG. This suggests the involvement of mTORC1 in the deleterious effect of ethanol on the developing brain.
Full article
►▼
Show Figures
Open AccessReview
TOR Signaling Pathway in Cardiac Aging and Heart Failure
by
Nastaran Daneshgar, Peter S. Rabinovitch and Dao-Fu Dai
Cited by 16 | Viewed by 4125
Abstract
Mechanistic Target of Rapamycin (mTOR) signaling is a key regulator of cellular metabolism, integrating nutrient sensing with cell growth. Over the past two decades, studies on the mTOR pathway have revealed that mTOR complex 1 controls life span, health span, and aging by
[...] Read more.
Mechanistic Target of Rapamycin (mTOR) signaling is a key regulator of cellular metabolism, integrating nutrient sensing with cell growth. Over the past two decades, studies on the mTOR pathway have revealed that mTOR complex 1 controls life span, health span, and aging by modulating key cellular processes such as protein synthesis, autophagy, and mitochondrial function, mainly through its downstream substrates. Thus, the mTOR pathway regulates both physiological and pathological processes in the heart from embryonic cardiovascular development to maintenance of cardiac homeostasis in postnatal life. In this regard, the dysregulation of mTOR signaling has been linked to many age-related pathologies, including heart failure and age-related cardiac dysfunction. In this review, we highlight recent advances of the impact of mTOR complex 1 pathway and its regulators on aging and, more specifically, cardiac aging and heart failure.
Full article
►▼
Show Figures
Planned Papers
The below list represents only planned manuscripts. Some of these
manuscripts have not been received by the Editorial Office yet. Papers
submitted to MDPI journals are subject to peer-review.
Title: Molecular mechanism underlying the kinase- independent neurite outgrowth by DGKbeta
Authors: Takuya Kano; Ryousuke Tsumagari; Nakasima; Ushio Kikkawa; Shuji Ueda; Minoru Yamanoue; Yasuhito Shirai
Affiliation: Department of Applied Chemistry in Bioscience, Graduate School of Agricultural Sciences & Faculty of Agriculture, Kobe University, Kobe, Japan
Title: To be determined
Authors: Ted Powers; et al.
Affiliation: Department of Molecular and Cellular Biology, University of California, Davis, CA 95616, USA
Title: Fission yeast TORC2 signaling
Authors: Shigeaki Saitoh; et al.
Affiliation: Department of Cell Biology, Institute of Life Science, Kurume University, Asahi-machi 67, Kurume, Fukuoka 830-0011, Japan
Title: To be determined
Authors: Madhulika B Gupta; et al.
Affiliation: Department of Biochemistry, University of Western Ontario, London, Ontario, Canada
Title: To be determined
Authors: Jacob Brunkard; et al.
Affiliation: University of Wisconsin-Madisondisabled, Madison, USA
Title: To be determined
Authors: Riko Hatakeyama; et al.
Affiliation: Institute of Medical Sciences, University of Aberdeen, Foresterhill, UK