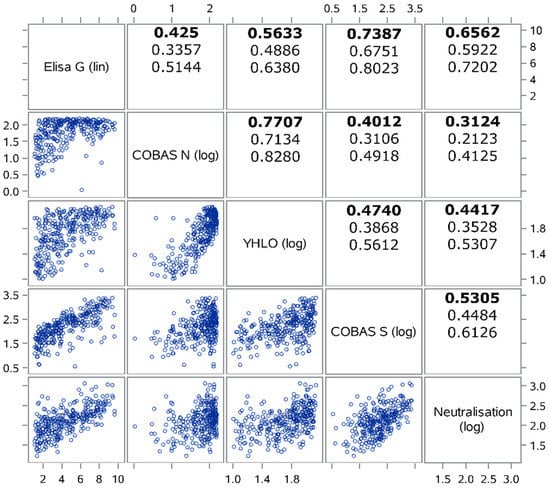
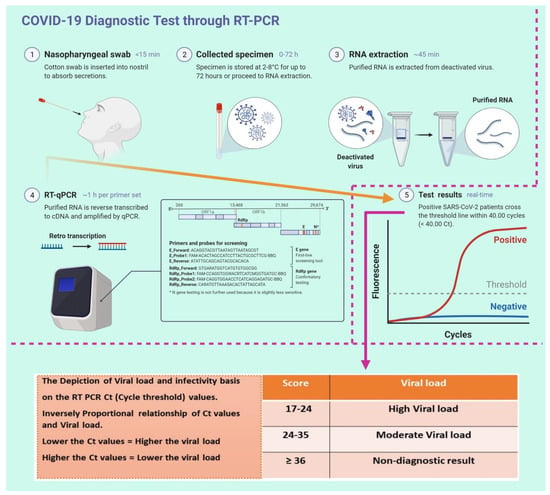
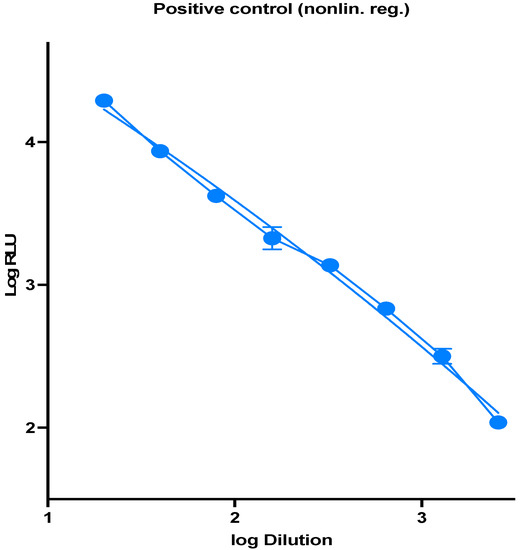
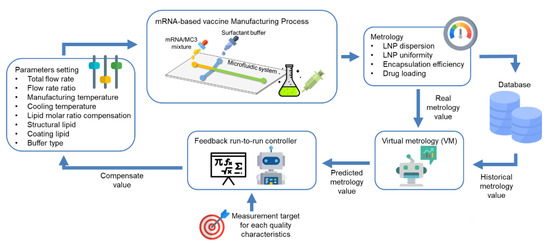
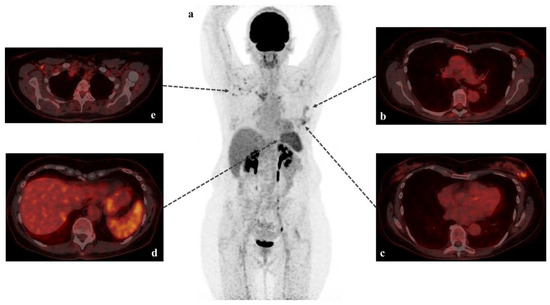
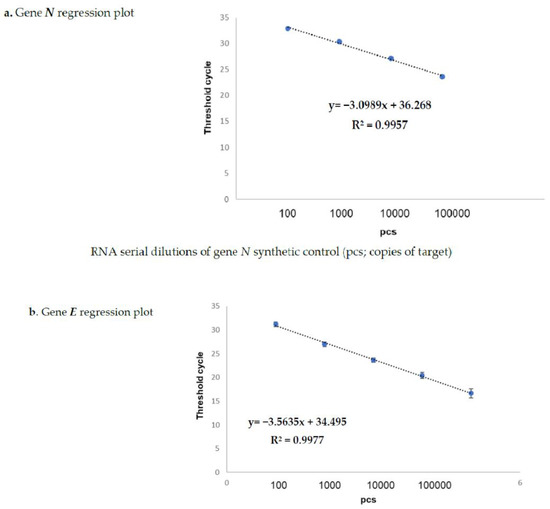
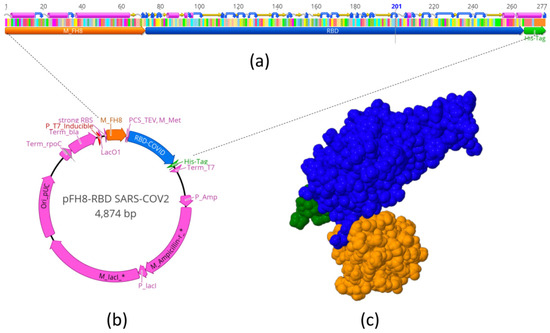
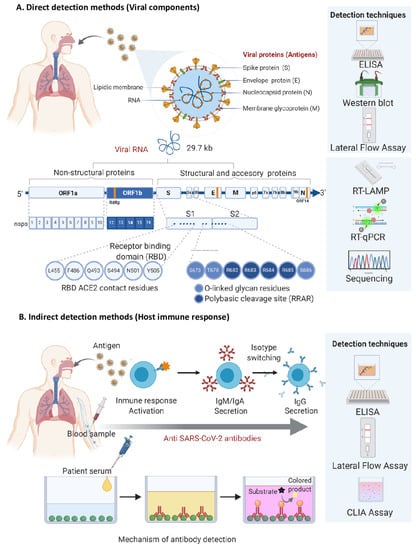
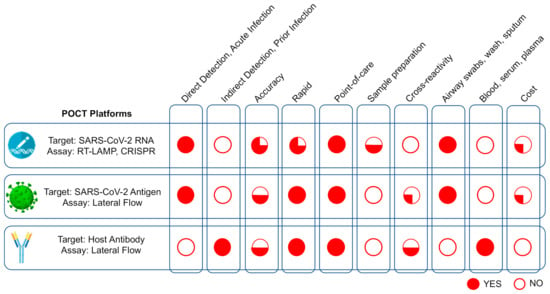
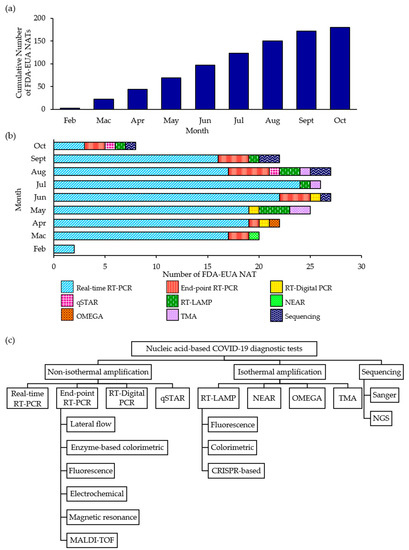
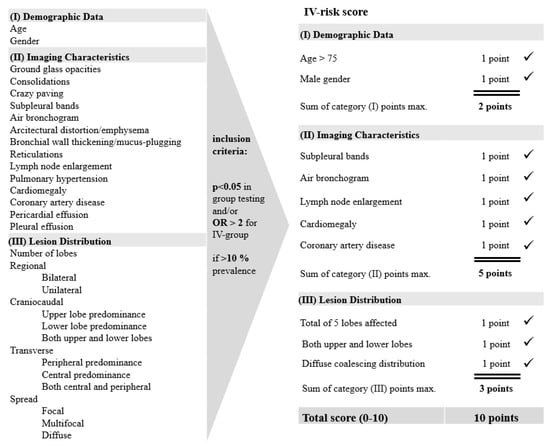
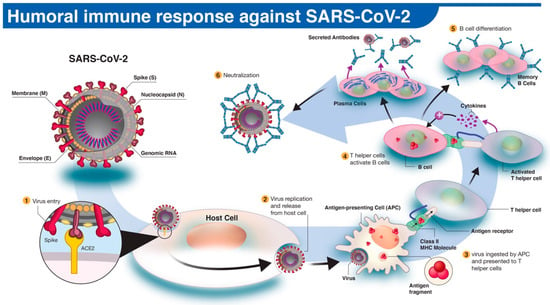
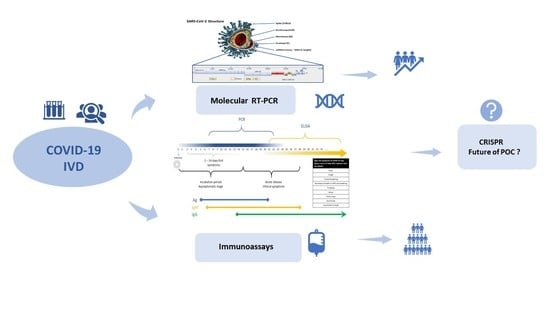
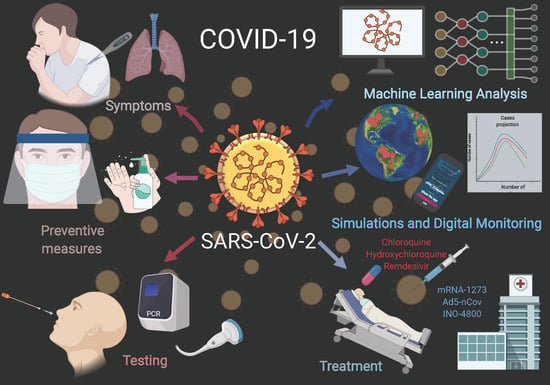
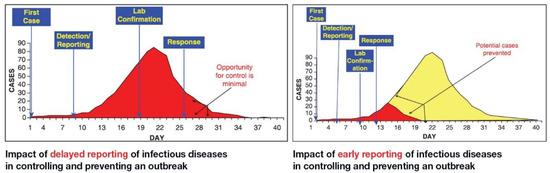
Virus Diagnostic Methods and Techniques: Learning from the COVID-19 Global Outbreak
A topical collection in Diagnostics (ISSN 2075-4418). This collection belongs to the section "Diagnostic Microbiology and Infectious Disease".
Viewed by 400766Editors
Interests: biomedical engineering; diagnostic methods; vaccines
Special Issues, Collections and Topics in MDPI journals
Interests: immunoassays; IVD; POC technologies; molecular diagnostics; next-generation sequencing; mobile healthcare
Special Issues, Collections and Topics in MDPI journals
Interests: human retroviruses (HIV-1, HIV-2 & HTLV); viral hepatitis (HBV, HCV, and HDV); resistance to antiretrovirals; molecular epidemiology for HTLV-1, HIV-2 subtypes
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
The spread of COVID-19 has become a global healthcare issue around the world. With recent technological advances in multiple research fields, such as materials science, micro-/nanotechnology, molecular biology, and bioengineering, much attention is shifting toward the development of new virus-based detection tools that not only address the needs for high sensitivity and specificity but also fulfil economic objectives in addition to the need for rapid point-of-care for groups and individuals with constrained resources and, possibly, limited training. These new detection technologies are also potentially applicable to different healthcare issues since they are disposable, inexpensive, portable, and easy to use—especially when their manufacture is based on low-cost materials. The topics in this Special Issue would cover point-of-care detection devices, microfluidic or paper-based detection devices, new materials for making detection devices, and others, with a particular focus on the precise diagnosis of COVID-19.
Dr. Chao-Min Cheng
Dr. Sandeep K. Vashist
Prof. Dr. Carmen de Mendoza
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Diagnostics is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
COVID-19
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
"Technologies for COVID-19 Diagnosis and Pandemic Response" Planned Book Chapter:
|
Chapters |
Title (MDPI Publication in COVID-19 Special Issue) |
Authors |
|
1. |
SARS-CoV-2 infection & pathophysiology |
Recruiting |
|
2. |
Role of diagnostics in the COVID-19 pandemic response |
Dr. Sandeep Kumar Vashist |
|
3. |
Biomarkers during COVID-19: Mechanisms of Change and Implications for Patient Outcomes |
Dr. Chao-Min Cheng |
|
4. |
In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10040202) |
Sandeep Kumar Vashist |
|
5. |
Nucleic Acid-Based Diagnostic Tests for the Detection SARS-CoV-2: An Update (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11010053) |
Choo Yee Yu, Kok Gan Chan, Chan Yean Yean & Geik Yong Ang
|
|
6. |
Viral Dynamics and Real-Time RT-PCR Ct Values Correlation with Disease Severity in COVID-19 (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11061091) |
Ali A. Rabaan, Raghavendra Tirupathi, Anupam A Sule, Jehad Aldali, Abbas Al Mutair, Saad Alhumaid, Muzaheed, Nitin Gupta, Thoyaja Koritala, Ramesh Adhikari, Muhammad Bilal, Manish Dhawan, Ruchi Tiwari, Saikat Mitra, Talha Bin Emran & Kuldeep Dhama |
|
7. |
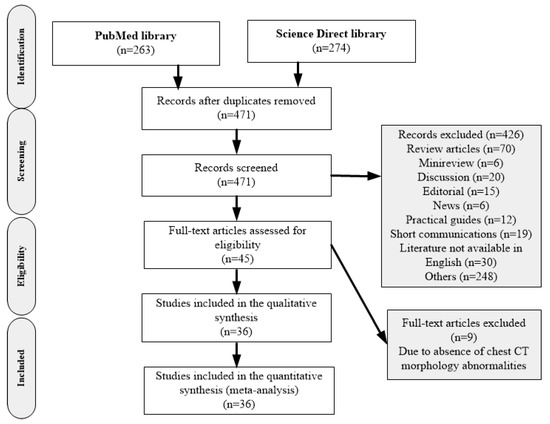
Prevalence of COVID-19 Diagnostic Output with Chest Computed Tomography: A Systematic Review and Meta-Analysis (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10121023) |
Temitope Emmanuel Komolafe, John Agbo, Ebenezer Obaloluwa Olaniyi, Kayode Komolafe |
|
8. |
COVID-19 Point-of-Care Diagnostics That Satisfy Global Target Product Profiles (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11010115) |
Abdi Ghaffari, Robyn Meurant & Ali Ardakani
|
|
9. |
Antibody Tests in Detecting SARS-CoV-2 Infection: A Meta-Analysis (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10050319) |
Panagiota I. Kontou, Georgia G. Braliou, Niki L. Dimou, Georgios Nikolopoulos & Pantelis G. Bagos |
|
10. |
COVID-19 Serological Tests: How Well Do They Actually Perform? (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10070453) |
Abdi Ghaffari, Robyn Meurant & Ali Ardakani |
|
11. |
Molecular and Serological Tests for COVID-19. A Comparative Review of SARS-CoV-2 Coronavirus Laboratory and Point-of-Care Diagnostics (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10060434) |
Robert Kubina & Arkadiusz Dziedzic
|
|
12. |
COVID-19 Diagnostics, Tools, and Prevention (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10060409) |
Mayar Allam, Shuangyi Cai, Shambavi Ganesh, Mythreye Venkatesan, Saurabh Doodhwala, Zexing Song, Thomas Hu, Aditi Kumar, Jeremy Heit, COVID-19 Study Group & Ahmet F. Coskun |
|
13. |
COVID-19 Infection Diagnosis: Potential Impact of Isothermal Amplification Technology to Reduce Community Transmission of SARS-CoV-2 (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10060399) |
Ameh S. James & John I. Alawneh
|
|
14. |
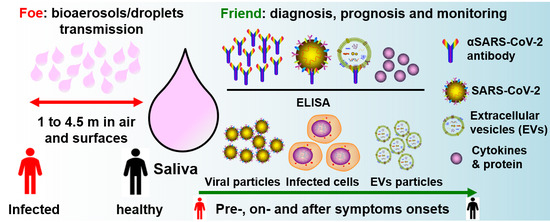
Saliva—Friend and Foe in the COVID-19 Outbreak (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10050290) |
Pingping Han & Sašo Ivanovski
|
|
15. |
Combining Point-of-Care Diagnostics and Internet of Medical Things (IoMT) to Combat the COVID-19 Pandemic (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics10040224) |
Ting Yang, Mattia Gentile, Ching-Fen Shen & Chao-Min Cheng
|
|
16. |
Electrochemical biosensors for COVID-19 |
Recruiting |
|
17. |
How Smart Manufacturing Can Help Combat the COVID-19 Pandemic (https://0-doi-org.brum.beds.ac.uk/10.3390/diagnostics11050885) |
Yun-Siang Lin, Chao-Min Cheng & Chen-Fu Chien
|
|
18. |
COVID-19 self-tests and their role in pandemic response |
Dr. Sandeep Kumar Vashist |
|
19. |
Role of mobile healthcare technologies, digital healthcare, artificial intelligence and telemedicine in COVID-19 pandemic response |
Recruiting |
|
20. |
Impact of COVID-19 on IVD industry and healthcare and how it will influence future pandemic response |
Recruiting |