Non-Coding RNA Methods
A topical collection in Non-Coding RNA (ISSN 2311-553X).
Viewed by 27337
Share This Topical Collection
Editors
 Dr. Piero Carninci
Dr. Piero Carninci
 Dr. Piero Carninci
Dr. Piero Carninci
E-Mail
Collection Editor
Laboratory for Transcriptome Technology, RIKEN Center for Integrative Medical Sciences, Yokohama, Kanagawa 230-0045, Japan
 Dr. Florent Hubé
Dr. Florent Hubé
 Dr. Florent Hubé
Dr. Florent Hubé
E-Mail
Website
Collection Editor
Transgenerational Epigenetics & Small RNA Biology, Sorbonne Université, CNRS, Laboratoire Biologie du Développement, Institut de Biologie Paris-Seine, UMR7622, 75005 Paris, France
Interests: long non-coding RNAs; small non-coding RNAs; functional genomics; transcriptional regulation; RNA secondary structure; RNA–protein interactions
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Non-coding RNA is now accepting submissions for a Topical Collection on non-coding RNA methods. This Topical Collection is co guest-edited by Dr. Piero Carninci from the RIKEN Center for Life Science Technologies in Japan and Dr. Florent Hubé; from the National Center for Scientific Research (CNRS) in Paris. Accepted papers are published online immediately after copy editing.
ncRNA is a rapidly growing field in which new research methods are continuously appearing. With this Topical Collection, we aim to collect a selection of articles on newly developed techniques of special interest for researchers in the ncRNA field.
We will consider manuscripts that report on new experimental and computational methods, as well as reviews of exceptional interest that focus on newly developed techniques for ncRNA research that include:
-
ncRNA analysis programs
-
ncRNA chromatography methods
-
ncRNA structure analysis
-
ncRNA function analysis
-
ncRNA by high-throughput screening
-
ncRNA database/annotation
-
ncRNA imaging
-
methods to detect the interactome of RNA (with proteins, with chromatin, etc.)
Dr. Piero Carninci
Dr. Florent Hubé
Collection Editors
Please use the online submission system and indicate in your cover letter that you would like to have your manuscript considered for the Topical Collection "Non Coding RNA methods". If you would like to enquire about the suitability of your article for this Topical Collection, please email your pre-submission enquiry to Dr. Piero Carninci ([email protected]) or to Dr. Florent Hubé ([email protected]) with the ncRNA Editorial Office ([email protected]) in copy.
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Non-Coding RNA is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 1800 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Published Papers (6 papers)
Open AccessProtocol
A Tool to Design Bridging Oligos Used to Detect Pseudouridylation Sites on RNA after CMC Treatment
by
Baptiste Bogard, Gilles Tellier, Claire Francastel and Florent Hubé
Viewed by 2125
Abstract
Pseudouridylation is one of the most abundant modifications found in RNAs. To identify the Pseudouridylation sites (Psi) in RNAs, several techniques have been developed, but the most common and robust is the CMC (N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide) treatment, which consists in the addition of an adduct
[...] Read more.
Pseudouridylation is one of the most abundant modifications found in RNAs. To identify the Pseudouridylation sites (Psi) in RNAs, several techniques have been developed, but the most common and robust is the CMC (N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide) treatment, which consists in the addition of an adduct on Psi that inhibits the reverse transcription. Here, we describe a turnkey method and a tool to design the bridging oligo (DBO), which is somewhat difficult to design. Finally, we propose a trouble-shooting guide to help users.
Full article
►▼
Show Figures
Open AccessArticle
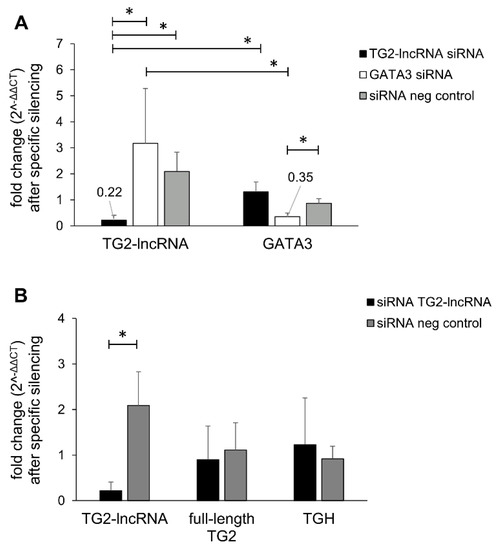
Inhibition of the lncRNA Coded within Transglutaminase 2 Gene Impacts Several Relevant Networks in MCF-7 Breast Cancer Cells
by
Carlo M. Bergamini, Chiara Vischioni, Gianluca Aguiari, Carmen Grandi, Anna Terrazzan, Stefano Volinia, Nicoletta Bianchi and Cristian Taccioli
Cited by 3 | Viewed by 3814
Abstract
Long non-coding RNAs are nucleotide molecules that regulate transcription in numerous cellular processes and are related to the occurrence of many diseases, including cancer. In this regard, we recently discovered a polyadenylated long non-coding RNA (named TG2-lncRNA) encoded within the first intron of
[...] Read more.
Long non-coding RNAs are nucleotide molecules that regulate transcription in numerous cellular processes and are related to the occurrence of many diseases, including cancer. In this regard, we recently discovered a polyadenylated long non-coding RNA (named TG2-lncRNA) encoded within the first intron of the Transglutaminase type 2 gene (
TGM2), which is related to tumour proliferation in human cancer cell lines. To better characterize this new biological player, we investigated the effects of its suppression in MCF-7 breast cancer cells, using siRNA treatment and RNA-sequencing. In this way, we found modifications in several networks associated to biological functions relevant for tumorigenesis (apoptosis, chronic inflammation, angiogenesis, immunomodulation, cell mobility, and epithelial–mesenchymal transition) that were originally attributed only to Transglutaminase type 2 protein but that could be regulated also by TG2-lncRNA. Moreover, our experiments strongly suggest the ability of TG2-lncRNA to directly interact with important transcription factors, such as RXRα and TP53, paving the way for several regulatory loops that can potentially influence the phenotypic behaviour of MCF-7 cells. These considerations imply the need to further investigate the relative relevance of the TG2 protein itself and/or other gene products as key regulators in the organization of breast cancer program.
Full article
►▼
Show Figures
Open AccessArticle
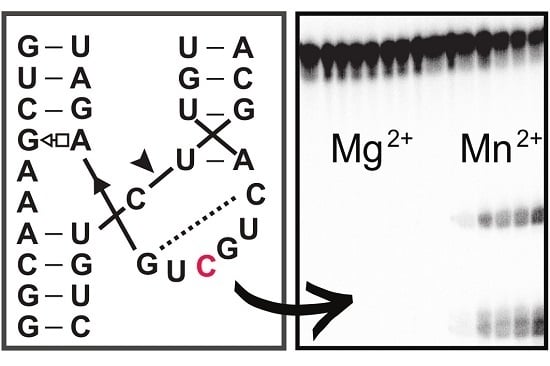
Single Mutation in Hammerhead Ribozyme Favors Cleavage Activity with Manganese over Magnesium
by
Mohammad Reza Naghdi, Emilie Boutet, Clarisse Mucha, Jonathan Ouellet and Jonathan Perreault
Cited by 1 | Viewed by 4784
Abstract
Hammerhead ribozymes are one of the most studied classes of ribozymes so far, from both the structural and biochemical point of views. The activity of most hammerhead ribozymes is cation-dependent. Mg
2+ is one of the most abundant divalent cations in the cell
[...] Read more.
Hammerhead ribozymes are one of the most studied classes of ribozymes so far, from both the structural and biochemical point of views. The activity of most hammerhead ribozymes is cation-dependent. Mg
2+ is one of the most abundant divalent cations in the cell and therefore plays a major role in cleavage activity for most hammerhead ribozymes. Besides Mg
2+, cleavage can also occur in the presence of other cations such as Mn
2+. The catalytic core of hammerhead ribozymes is highly conserved, which could contribute to a preference of hammerhead ribozymes toward certain cations. Here, we show a naturally occurring variation in the catalytic core of hammerhead ribozymes, A6C, that can favor one metallic ion, Mn
2+, over several other cations.
Full article
►▼
Show Figures
Open AccessFeature PaperArticle
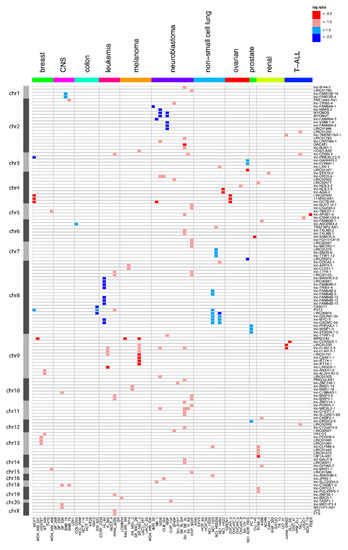
Targeted Genomic Screen Reveals Focal Long Non-Coding RNA Copy Number Alterations in Cancer Cell Lines
by
Pieter-Jan Volders, Steve Lefever, Shalina Baute, Justine Nuytens, Katrien Vanderheyden, Björn Menten, Pieter Mestdagh and Jo Vandesompele
Cited by 6 | Viewed by 3815
Abstract
The landscape of somatic copy-number alterations (SCNAs) affecting long non-coding RNAs (lncRNAs) in human cancers remains largely unexplored. While the majority of lncRNAs remain to be functionally characterized, several have been implicated in cancer development and metastasis. Considering the plethora of lncRNAs genes
[...] Read more.
The landscape of somatic copy-number alterations (SCNAs) affecting long non-coding RNAs (lncRNAs) in human cancers remains largely unexplored. While the majority of lncRNAs remain to be functionally characterized, several have been implicated in cancer development and metastasis. Considering the plethora of lncRNAs genes that have been currently reported, it is conceivable that many more lncRNAs might function as oncogenes or tumor suppressor genes. We devised a strategy to detect focal lncRNA SCNAs using a custom DNA microarray platform probing 10,519 lncRNA genes. By screening a panel of 80 cancer cell lines, we detected numerous focal aberrations targeting one or multiple lncRNAs without affecting neighboring protein-coding genes. These focal aberrations are highly suggestive for a tumor suppressive or oncogenic role of the targeted lncRNA gene. Although functional validation remains an essential step in the further characterization of the involved candidate cancer lncRNAs, our results provide a direct way of prioritizing candidate lncRNAs that are involved in cancer pathogenesis.
Full article
►▼
Show Figures
Open AccessArticle
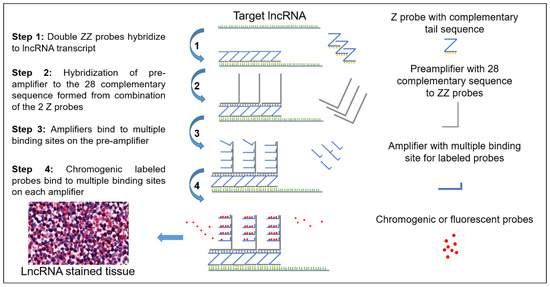
Z Probe, An Efficient Tool for Characterizing Long Non-Coding RNA in FFPE Tissues
by
Manish K. Tripathi, Chidi Zacheaus, Kyle Doxtater, Fatemeh Keramatnia, Cuilan Gao, Murali M. Yallapu, Meena Jaggi and Subhash C. Chauhan
Cited by 8 | Viewed by 5638
Abstract
Formalin-fixed paraffin embedded (FFPE) tissues are a valuable resource for biomarker discovery in order to understand the etiology of different cancers and many other diseases. Proteins are the biomarkers of interest with respect to FFPE tissues as RNA degradation is the major challenge
[...] Read more.
Formalin-fixed paraffin embedded (FFPE) tissues are a valuable resource for biomarker discovery in order to understand the etiology of different cancers and many other diseases. Proteins are the biomarkers of interest with respect to FFPE tissues as RNA degradation is the major challenge in these tissue samples. Recently, non-protein coding transcripts, long non-coding RNAs (lncRNAs), have gained significant attention due to their important biological actions and potential involvement in cancer. RNA sequencing (RNA-seq) or quantitative reverse transcription-polymerase chain reaction (qRT-PCR) are the only validated methods to evaluate and study lncRNA expression and neither of them provides visual representation as immunohistochemistry (IHC) provides for proteins. We have standardized and are reporting a sensitive Z probe based in situ hybridization method to visually identify and quantify lncRNA in FFPE tissues. This assay is highly sensitive and identifies transcripts visible within different cell types and tumors. We have detected a scarcely expressed tumor suppressor lncRNA NRON (non-coding repressor of nuclear factor of activated T-cells (NFAT)), a moderately expressed oncogenic lncRNA UCA1 (urothelial cancer associated 1), and a highly studied and expressed lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1) in different cancers. High MALAT1 staining was found in colorectal, breast and pancreatic cancer. Additionally, we have observed an increase in MALAT1 expression in different stages of colorectal cancer.
Full article
►▼
Show Figures
Open AccessCommunication
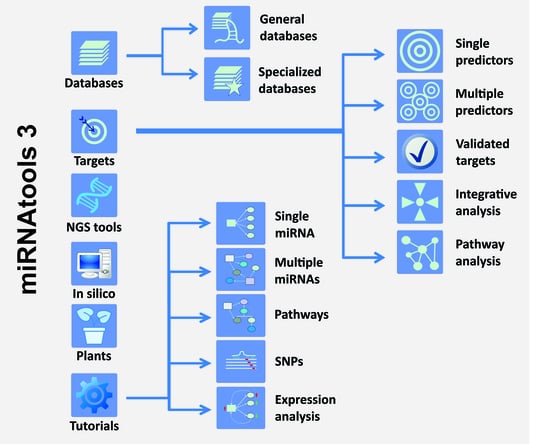
miRNAtools: Advanced Training Using the miRNA Web of Knowledge
by
Ewa Ł. Stępień, Marina C. Costa and Francisco J. Enguita
Cited by 4 | Viewed by 5418
Abstract
Micro-RNAs (miRNAs) are small non-coding RNAs that act as negative regulators of the genomic output. Their intrinsic importance within cell biology and human disease is well known. Their mechanism of action based on the base pairing binding to their cognate targets have helped
[...] Read more.
Micro-RNAs (miRNAs) are small non-coding RNAs that act as negative regulators of the genomic output. Their intrinsic importance within cell biology and human disease is well known. Their mechanism of action based on the base pairing binding to their cognate targets have helped the development not only of many computer applications for the prediction of miRNA target recognition but also of specific applications for functional assessment and analysis. Learning about miRNA function requires practical training in the use of specific computer and web-based applications that are complementary to wet-lab studies. In order to guide the learning process about miRNAs, we have created miRNAtools (
http://mirnatools.eu), a web repository of miRNA tools and tutorials. This article compiles tools with which miRNAs and their regulatory action can be analyzed and that function to collect and organize information dispersed on the web. The miRNAtools website contains a collection of tutorials that can be used by students and tutors engaged in advanced training courses. The tutorials engage in analyses of the functions of selected miRNAs, starting with their nomenclature and genomic localization and finishing with their involvement in specific cellular functions.
Full article
►▼
Show Figures