The Development of Assays for Heparanase Enzymatic Activity: Towards a Gold Standard
Abstract
:1. Introduction
2. Heparanase Mechanism and Substrate Specificity
3. Assaying Heparanase
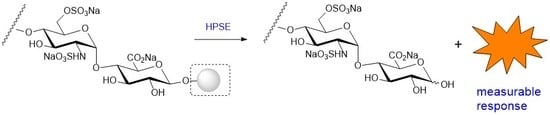
3.1. Heparin/HS-Based Assays
3.2. Homogeneous HS Oligosaccharide Substrates
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sanderson, R.D.; Elkin, M.; Rapraeger, A.C.; Ilan, N.; Vlodavsky, I. Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 2017, 284, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Singh, P.; Boyango, I.; Gutter-Kapon, L.; Elkin, M.; Sanderson, R.D.; Ilan, N. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist. Updat. 2016, 29, 54–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.P.; Kusche-Gullberg, M. Heparan sulfate: Biosynthesis, structure, and function. Int. Rev. Cell Mol. Biol. 2016, 325, 215–273. [Google Scholar] [PubMed]
- Rivara, S.; Milazzo, F.M.; Giannini, G. Heparanase: A rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Med. Chem. 2016, 8, 647–680. [Google Scholar] [CrossRef] [PubMed]
- Simeonovic, C.J.; Ziolkowski, A.F.; Wu, Z.; Choong, F.J.; Freeman, C.; Parish, C.R. Heparanase and autoimmune diabetes. Front. Immunol. 2013, 4, 471. [Google Scholar] [CrossRef] [PubMed]
- Rabelink, T.J.; van den Berg, B.M.; Garsen, M.; Wang, G.; Elkin, M.; van der Vlag, J. Heparanase: Roles in cell survival, extracellular matrix remodelling and the development of kidney disease. Nat. Rev. Nephrol. 2017, 13, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Blich, M.; Li, J.P.; Sanderson, R.D.; Ilan, N. Involvement of heparanase in atherosclerosis and other vessel wall pathologies. Matrix Biol. 2013, 32, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, N.; Yadavalli, T.; Jaishankar, D.; Shukla, D. Emerging Roles of Heparanase in Viral Pathogenesis. Pathogens 2017, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Irimura, T.; Di Ferrante, D.; Di Ferrante, N.; Nicolson, G.L. Heparan sulfate degradation: Relation to tumor invasive and metastatic properties of mouse B16 melanoma sublines. Science 1983, 220, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Fuks, Z.; Bar-Ner, M.; Ariav, Y.; Schirrmacher, V. Lymphoma cell-mediated degradation of sulfated proteoglycans in the subendothelial extracellular matrix: Relationship to tumor cell metastasis. Cancer Res. 1983, 43, 2704–2711. [Google Scholar] [PubMed]
- Hulett, M.D.; Freeman, C.; Hamdorf, B.J.; Baker, R.T.; Harris, M.J.; Parish, C.R. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat. Med. 1999, 5, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Friedmann, Y.; Elkin, M.; Aingorn, H.; Atzmon, R.; Ishai-Michaeli, R.; Bitan, M.; Pappo, O.; Peretz, T.; Michal, I.; et al. Mammalian heparanase: Gene cloning, expression and function in tumor progression and metastasis. Nat. Med. 1999, 5, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Viola, C.M.; Brzozowski, A.M.; Davies, G.J. Structural characterization of human heparanase reveals insights into substrate recognition. Nat. Struct. Mol. Biol. 2015, 22, 1016–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, J.C.; Laloo, A.E.; Singh, S.; Ferro, V. 1H-NMR spectroscopic studies establish that heparanase is a retaining glycosidase. Biochem. Biophys. Res. Commun. 2014, 443, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Pikas, D.S.; Li, J.P.; Vlodavsky, I.; Lindahl, U. Substrate specificity of heparanases from human hepatoma and platelets. J. Biol. Chem. 1998, 273, 18770–18777. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Yamada, S.; Toyoshima, M.; Dong, J.; Nakajima, M.; Sugahara, K. Structural recognition by recombinant human heparanase that plays critical roles in tumor metastasis. Hierarchical sulfate groups with different effects and the essential target disulfated trisaccharide sequence. J. Biol. Chem. 2002, 277, 42488–42495. [Google Scholar] [CrossRef] [PubMed]
- Bisio, A.; Mantegazza, A.; Urso, E.; Naggi, A.; Torri, G.; Viskov, C.; Casu, B. High-performance liquid chromatographic/mass spectrometric studies on the susceptibility of heparin species to cleavage by heparanase. Semin. Thromb. Hemost. 2007, 33, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.B.; Liu, J. Unraveling the specificity of heparanase utilizing synthetic substrates. J. Biol. Chem. 2010, 285, 14504–14513. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.; Liu, J. Deciphering mode of action of heparanase using structurally defined oligosaccharides. J. Biol. Chem. 2012, 287, 34836–34843. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Huang, Y.; Buczek-Thomas, J.A.; Ethen, C.M.; Nugent, M.A.; Wu, Z.L.; Zaia, J. A liquid chromatography-mass spectrometry-based approach to characterize the substrate specificity of mammalian heparanase. J. Biol. Chem. 2014, 289, 34141–34151. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.B.; Liu, J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013, 32, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Irimura, T.; Nicolson, G.L. A solid-phase substrate of heparanase: Its application to assay of human melanoma for heparan sulfate degradative activity. Anal. Biochem. 1986, 157, 162–171. [Google Scholar] [CrossRef]
- Xu, Y.J.; Miao, H.Q.; Pan, W.; Navarro, E.C.; Tonra, J.R.; Mitelman, S.; Camara, M.M.; Deevi, D.S.; Kiselyov, A.S.; Kussie, P.; et al. N-(4-{[4-(1H-Benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamide derivatives as small molecule heparanase inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Nardella, C.; Steinkuhler, C. Radiolabeled heparan sulfate immobilized on microplate as substrate for the detection of heparanase activity. Anal. Biochem. 2004, 332, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.; Parish, C.R. A rapid quantitative assay for the detection of mammalian heparanase activity. Biochem. J. 1997, 325, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-Ner, M.; Eldor, A.; Wasserman, L.; Matzner, Y.; Cohen, I.R.; Fuks, Z.; Vlodavsky, I. Inhibition of heparanase-mediated degradation of extracellular matrix heparan sulfate by non-anticoagulant heparin species. Blood 1987, 70, 551–557. [Google Scholar] [PubMed]
- Nakajima, M.; DeChavigny, A.; Johnson, C.E.; Hamada, J.; Stein, C.A.; Nicolson, G.L. Suramin. A potent inhibitor of melanoma heparanase and invasion. J. Biol. Chem. 1991, 266, 9661–9666. [Google Scholar] [PubMed]
- Lapierre, F.; Holme, K.; Lam, L.; Tressler, R.J.; Storm, N.; Wee, J.; Stack, R.J.; Castellot, J.; Tyrrell, D.J. Chemical modifications of heparin that diminish its anticoagulant but preserve its heparanase-inhibitory, angiostatic, anti-tumor and anti- metastatic properties. Glycobiology 1996, 6, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Podyma-Inoue, K.A.; Yanagishita, M. Ultrafiltration-based assay for heparanase activity. Anal. Biochem. 2004, 331, 147–152. [Google Scholar] [CrossRef]
- Karoli, T.; Liu, L.; Fairweather, J.K.; Hammond, E.; Li, C.P.; Cochran, S.; Bergefall, K.; Trybala, E.; Addison, R.S.; Ferro, V. Synthesis, Biological Activity, and Preliminary Pharmacokinetic Evaluation of Analogues of a Phosphosulfomannan Angiogenesis Inhibitor (PI-88). J. Med. Chem. 2005, 48, 8229–8236. [Google Scholar] [CrossRef] [PubMed]
- Fairweather, J.K.; Hammond, E.; Johnstone, K.D.; Ferro, V. Synthesis and heparanase inhibitory activity of sulfated mannooligosaccharides related to the antiangiogenic agent PI-88. Bioorg. Med. Chem. 2008, 16, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Bame, K.J.; Danda, J.; Hassall, A.; Tumova, S. Aβ(1-40) prevents heparanase-catalyzed degradation of heparan sulfate glycosaminoglycans and proteoglycans in vitro. A role for heparan sulfate proteoglycan turnover in Alzheimer’s disease. J. Biol. Chem. 1997, 272, 17005–17011. [Google Scholar] [CrossRef] [PubMed]
- De Vouge, M.W.; Yamazaki, A.; Bennett, S.A.; Chen, J.H.; Shwed, P.S.; Couture, C.; Birnboim, H.C. Immunoselection of GRP94/endoplasmin from a KNRK cell-specific lambda gt11 library using antibodies directed against a putative heparanase amino-terminal peptide. Int. J. Cancer 1994, 56, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Teruya, T.; Simizu, S.; Osada, H. Exploitation of heparanase inhibitors from microbial metabolites using an efficient visual screening system. J. Antibiot. 2004, 57, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Ben-Artzi, H.; Ayal-Hershkovitz, M.; Vlodavsky, I.; Pecker, I.; Peleg, Y.; Miron, D. Method of Screening for Potential Anti-Metastatic and Anti-Inflammatory Agents Using Mammalian Heparanase as a Probe. US Patent US 6,190,875 B1, 20 February 2001. [Google Scholar]
- Behzad, F.; Brenchley, P.E. A multiwell format assay for heparanase. Anal. Biochem. 2003, 320, 207–213. [Google Scholar] [CrossRef]
- Huang, K.S.; Holmgren, J.; Reik, L.; Lucas-McGady, D.; Roberts, J.; Liu, C.M.; Levin, W. High-throughput methods for measuring heparanase activity and screening potential antimetastatic and anti-inflammatory agents. Anal. Biochem. 2004, 333, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, K.; Okamoto, H.; Numata, Y.; Takemoto, H. A simple and rapid assay for heparanase activity using homogeneous time-resolved fluorescence. J. Pharm. Biomed. Anal. 2006, 41, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Cisbio. Heparanase Assay Toolbox. Available online: https://www.cisbio.com/drug-discovery/heparanase-assay-toolbox (accessed on 30 October 2018).
- Sistla, J.C.; Morla, S.; Alabbas, A.-H.B.; Kalathur, R.C.; Sharon, C.; Patel, B.B.; Desai, U.R. Polymeric fluorescent heparin as one-step FRET substrate of human heparanase. Carbohydr. Polym. 2019, 205, 385–391. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Shi, C.; Wu, X.R.; Chen, Y.J. Efficient access to the non-reducing end of low molecular weight heparin for fluorescent labeling. Chem. Commun. 2014, 50, 7004–7006. [Google Scholar] [CrossRef] [PubMed]
- Melo, C.M.; Tersariol, I.L.; Nader, H.B.; Pinhal, M.A.; Lima, M.A. Development of new methods for determining the heparanase enzymatic activity. Carbohydr. Res. 2015, 412, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.L.; Huang, X.; Ethen, C.M.; Tatge, T.; Pasek, M.; Zaia, J. Non-reducing end labeling of heparan sulfate via click chemistry and a high throughput ELISA assay for heparanase. Glycobiology 2017, 27, 518–524. [Google Scholar] [CrossRef] [PubMed]
- TaKaRa Bio. Heparan Degrading Enzyme Assay Kit. Available online: https://www.takarabio.com (accessed on 30 October 2018).
- Haubeck, H.D. Method for Determining Endoglycosidase (Heparanase) Enzyme Activity. PCT Int. Appl. WO 2008/055575 A1, 15 May 2008. [Google Scholar]
- Courtney, S.M.; Hay, P.A.; Buck, R.T.; Colville, C.S.; Porter, D.W.; Scopes, D.I.C.; Pollard, F.C.; Page, M.J.; Bennett, J.M.; Hircock, M.L.; et al. 2,3-Dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid derivatives: A novel class of small molecule heparanase inhibitors. Bioorg. Med. Chem. Lett. 2004, 14, 3269–3273. [Google Scholar] [CrossRef] [PubMed]
- Galantos. Galantos Heparanase Assay. Available online: https://www.galantos.eu/heparanase/pdf/galantos_broschuere_hepa2010.pdf (accessed on 30 October 2108).
- Nasser, N.J.; Sarig, G.; Brenner, B.; Nevo, E.; Goldshmidt, O.; Zcharia, E.; Li, J.P.; Vlodavsky, I. Heparanase neutralizes the anticoagulation properties of heparin and low-molecular-weight heparin. J. Thromb. Haemost. 2006, 4, 560–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, S.C.; Kim, B.Y.; Oh, W.K.; Park, Y.M.; Kim, H.M.; Ahn, J.S. Colorimetric heparinase assay for alternative anti-metastatic activity. Life Sci. 2006, 79, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Ban, Z.; Bosques, C.J.; Sasisekharan, R. A simple assay to probe disease-associated enzyme activity using glycosaminoglycan-assisted synthesized gold nanoparticles. Org. Biomol. Chem. 2008, 6, 4290–4292. [Google Scholar] [CrossRef] [PubMed]
- Petitou, M.; van Boeckel, C.A. A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next? Angew. Chem. Int. Ed. Engl. 2004, 43, 3118–3133. [Google Scholar] [CrossRef] [PubMed]
- Driguez, P.A.; Petitou, M. Azasugar Derivatives, Heparanase Inhibitors, Method for Preparing Same, Compositions Containing Same, Use Thereof. PCT Int. Appl. WO 2006/021653 A2, 2 March 2006. [Google Scholar]
- Schiemann, S.; Luhn, S.; Alban, S. Development of both colorimetric and fluorescence heparinase activity assays using fondaparinux as substrate. Anal. Biochem. 2012, 427, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.; Li, C.P.; Ferro, V. Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Anal. Biochem. 2010, 396, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dredge, K.; Hammond, E.; Davis, K.; Li, C.P.; Liu, L.; Johnstone, K.; Handley, P.; Wimmer, N.; Gonda, T.J.; Gautam, A.; et al. The PG500 series: Novel heparan sulfate mimetics as potent angiogenesis and heparanase inhibitors for cancer therapy. Invest. New Drugs 2010, 28, 276–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferro, V.; Liu, L.; Johnstone, K.D.; Wimmer, N.; Karoli, T.; Handley, P.; Rowley, J.; Dredge, K.; Li, C.P.; Hammond, E.; et al. Discovery of PG545: A Highly Potent and Simultaneous Inhibitor of Angiogenesis, Tumor Growth, and Metastasis. J. Med. Chem. 2012, 55, 3804–3813. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.K.; Vierfuss, S.; Luhn, S.; Alban, S. Testing of potential glycan-based heparanase inhibitors in a fluorescence activity assay using either bacterial heparinase II or human heparanase. J. Pharm. Biomed. Anal. 2014, 95, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Petitou, M.; Duchaussoy, P.; Lederman, I.; Choay, J.; Sinay, P. Binding of heparin to antithrombin III: A chemical proof of the critical role played by a 3-sulfated 2-amino-2-deoxy-d-glucose residue. Carbohydr. Res. 1988, 179, 163–172. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, Y.; Chen, C.; Xu, W.; Yu, B. Synthesis of a tetrasaccharide substrate of heparanase. Carbohydr. Res. 2008, 343, 2853–2862. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Ikeda-Matsumi, R.; Ebara-Nagahara, K.; Otaki-Nanjo, M.; Taniguchi-Morita, K.; Nanjo, M.; Tamura, J. Synthesis of heparan sulfate tetrasaccharide as a substrate for human heparanase. Carbohydr. Res. 2012, 353, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Xu, W.; Dai, Y.; Yang, Y.; Yu, B. Efficient synthesis of a library of heparin tri- and tetrasaccharides relevant to the substrate of heparanase. Org. Chem. Front. 2014, 1, 405–414. [Google Scholar] [CrossRef]
- Pearson, A.G.; Kiefel, M.J.; Ferro, V.; von Itzstein, M. Synthesis of simple heparanase substrates. Org. Biomol. Chem. 2011, 9, 4614–4625. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, A.; Capo, A.; Caira, S.; Tramice, A.; Varriale, A.; Staiano, M.; D’Auria, S. Cloning and bacterial expression systems for recombinant human heparanase production: Substrate specificity investigation by docking of a putative heparanase substrate. Biotechnol. Appl. Biochem. 2018, 65, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Ohmae, M.; Fujita, Y.; Takada, J.; Kimura, S. Synthesis of a Heparan Sulfate Disaccharide Fluoride for Detection of Heparanase Activity. Chem. Lett. 2013, 42, 1168–1169. [Google Scholar] [CrossRef]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chhabra, M.; Ferro, V. The Development of Assays for Heparanase Enzymatic Activity: Towards a Gold Standard. Molecules 2018, 23, 2971. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23112971
Chhabra M, Ferro V. The Development of Assays for Heparanase Enzymatic Activity: Towards a Gold Standard. Molecules. 2018; 23(11):2971. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23112971
Chicago/Turabian StyleChhabra, Mohit, and Vito Ferro. 2018. "The Development of Assays for Heparanase Enzymatic Activity: Towards a Gold Standard" Molecules 23, no. 11: 2971. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23112971