The Characteristics of PD-L1 Inhibitors, from Peptides to Small Molecules
Abstract
:1. Introduction
2. Results
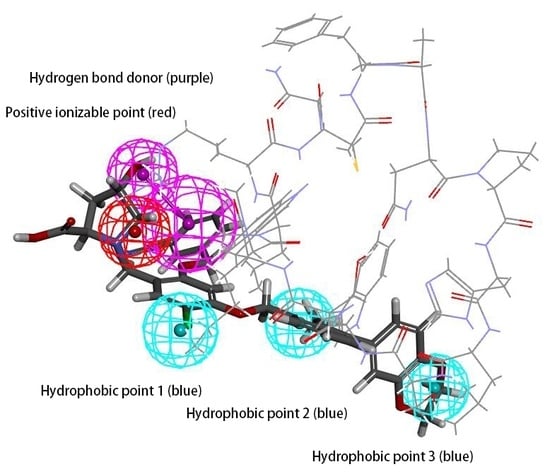
2.1. Pharmacophore Model Hypo 1A Hypotheses and Validation
2.2. Decoy Set of Hypo 1A
2.3. ROC Curve of Hypo 1A
2.4. Pharmacophore Model Hypo 1B Hypotheses and Validation
2.5. Decoy Set of Hypo 1B
2.6. ROC Curve of Hypo 1B
2.7. Molecular Docking Study
3. Materials and Methods
3.1. Generation and Validation of the Pharmacophore Model Based on Peptides
3.2. Generation and Validation of the Pharmacophore Model Based on Small Molecules
3.3. Molecular Docking Study
3.4. Analysing Interactions between PD-L1 and BMS Compounds
3.5. Comparing the Model Based on Peptides with the Model Based on Small Molecules
4. Discussion
4.1. Features of Hypo 1A
4.2. Features of Hypo 1B
4.3. Comparison between Hypo 1A and Hypo 1B
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Massari, F.; Santoni, M.; Ciccarese, C.; Daniele, S. The Immunocheckpoints in Modern Oncology: The next 15 Years. Expert Opin. Biol. Ther. 2015, 15, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Bellmunt, J.; Powles, T.; Vogelzang, N.J. A Review on the Evolution of Pd-1/Pd-L1 Immunotherapy for Bladder Cancer: The Future is Now. Cancer Treat. Rev. 2017, 54, 58–67. [Google Scholar] [CrossRef]
- Boussiotis, V.A. Molecular and Biochemical Aspects of The Pd-1 Checkpoint Pathway. New Engl. J. Med. 2016, 375, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Collin, M. Immune Checkpoint Inhibitors: A Patent Review (2010–2015). Expert Opin. Ther. Patents 2016, 26, 555–564. [Google Scholar] [CrossRef]
- Wajant, H. Death Receptors. Essays Biochem. 2003, 39, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Osthoff, K.; Ferrari, D.; Los, M.; Wesselborg, S.; Peter, M.E. Apoptosis Signaling by Death Receptors. Eur. J. Biochem. 1998, 254, 439–459. [Google Scholar] [CrossRef]
- Ahmed, M.; Barakat, K. The Too Many Faces of Pd-L1: A Comprehensive Conformational Analysis Study. Biochemistry 2017, 56, 5428–5439. [Google Scholar] [CrossRef] [PubMed]
- Dermani, F.K.; Samadi, P.; Rahmani, G.; Kohlan, A.K.; Najafi, R. Pd-1/Pd-L1 Immune Checkpoint: Potential Target for Cancer Therapy. J. Cell Physiol. 2019, 234, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of Pd-1 And Pd-L1 Inhibitors as A form of Cancer Immunotherapy: A Comprehensive Review of Registration Trials and Future Considerations. J. Immunother Cancer 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the Pd-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef]
- Khunger, M.; Rakshit, S.; Pasupuleti, V.; Hernandez, A.V.; Mazzone, P.; Stevenson, J.; Pennell, N.A.; Velcheti, V. Incidence of Pneumonitis with Use of Programmed Death 1 and Programmed Death-Ligand 1 Inhibitors in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis of Trials. Chest 2017, 152, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Kythreotou, A.; Siddique, A.; Mauri, F.A.; Bower, M.; Pinato, D.J. Pd-L1. J. Clin. Pathol. 2018, 71, 189–194. [Google Scholar] [CrossRef]
- Spagnuolo, A.; Gridelli, C. “Comparison of the Toxicity Profile of Pd-1 Versus Pd-L1 Inhibitors in Non-Small Cell Lung Cancer”: Is There a Substantial Difference or not? J. Thorac. Dis. 2018, 10, S4065. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, Q.; Liu, Z.; Chen, Y.; Feng, F.; Sun, H. Peptide-Based and Small Synthetic Molecule Inhibitors on Pd-1/Pd-L1 Pathway: A New Choice for Immunotherapy? Eur. J. Med. Chem. 2019, 161, 378–398. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.P.; Fink, M.A.; Enley, E.S.; Fisher, J.E.; Herb, M.C.; Klingos, A.; Proulx, J.T.; Fedorky, M.T. Identification of Small-Molecule Inhibitors of Pd-1/Pd-L1 Protein-Protein Interaction. Chemistryselect 2018, 3, 2185–2189. [Google Scholar] [CrossRef]
- Mondanelli, G.; Volpi, C.; Orabona, C.; Grohmann, U. Challenges in the Design of Reliable Immuno-Oncology Mouse Models to Inform Drug Development. Future Med. Chem. 2017, 9, 1313–1317. [Google Scholar] [CrossRef]
- Yang, J.; Hu, L. Immunomodulators Targeting The Pd-1/Pd-L1 Protein-Protein Interaction: From Antibodies To Small Molecules. Med. Res. Rev. 2019, 39, 265–301. [Google Scholar] [CrossRef]
- Sasikumar, P.G.; Ramachandra, M. Small-Molecule Antagonists of the Immune Checkpoint Pathways: Concept to Clinic. Future Med. Chem. 2017, 9, 1305–1308. [Google Scholar] [CrossRef]
- Powderly, J.; Patel, M.R.; Lee, J.J.; Brody, J.; Meric-Bernstam, F.; Hamilton, E.; Aix, S.P.; Garcia-Corbacho, J.; Bang, Y.J.; Ahn, M.J.; et al. Ca-170, A First in Class Oral Small Molecule Dual Inhibitor of Immune Checkpoints Pd-L1 and Vista, Demonstrates Tumor Growth Inhibition in Pre-Clinical Models and Promotes T Cell Activation in Phase 1 Study. Ann. Oncol. 2017, 28. [Google Scholar] [CrossRef]
- Li, K.; Tian, H. Development of Small-Molecule Immune Checkpoint Inhibitors of Pd-1/Pd-L1 as A New Therapeutic Strategy for Tumor Immunotherapy. J. Drug Target. 2019, 27, 244–256. [Google Scholar] [CrossRef]
- Kopalli, S.R.; Kang, T.B.; Lee, K.H.; Koppula, S. Novel Small Molecule Inhibitors of Programmed Cell Death (Pd)-1, And Its Ligand, Pd-L1 in Cancer Immunotherapy: A Review Update of Patent Literature. Recent Pat. Anticancer Drug Discov. 2018. [Google Scholar] [CrossRef]
- Cheng, B.; Yuan, W.E.; Su, J.; Liu, Y.; Chen, J. Recent Advances in Small Molecule Based Cancer Immunotherapy. Eur. J. Med. Chem. 2018, 157, 582–598. [Google Scholar] [CrossRef]
- Zak, K.M.; Kitel, R.; Przetocka, S.; Golik, P.; Guzik, K.; Musielak, B.; Domling, A.; Dubin, G.; Holak, T.A. Structure of the Complex of Human Programmed Death 1, Pd-1, and Its Ligand Pd-L1. Structure 2015, 23, 1348–2341. [Google Scholar] [CrossRef]
- Magiera-Mularz, K.; Skalniak, L.; Zak, K.M.; Musielak, B.; Rudzinska-Szostak, E.; Berlicki, L.; Kocik, J.; Grudnik, P.; Sala, D.; Zarganes-Tzitzikas, T.; et al. Bioactive Macrocyclic Inhibitors of the Pd-1/Pd-L1 Immune Checkpoint. Angew Chem. Int. Ed. Engl. 2017, 56, 13732–13735. [Google Scholar] [CrossRef]
- Zak, K.M.; Grudnik, P.; Guzik, K.; Zieba, B.J.; Musielak, B.; Domling, A.; Dubin, G.; Holak, T.A. Structural Basis For Small Molecule Targeting of The Programmed Death Ligand 1 (Pd-L1). Oncotarget 2016, 7, 30323–30335. [Google Scholar] [CrossRef]
- Guzik, K.; Zak, K.M.; Grudnik, P.; Magiera, K.; Musielak, B.; Torner, R.; Skalniak, L.; Domling, A.; Dubin, G.; Holak, T.A. Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (Pd-1/Pd-L1) Interaction via Transiently Induced Protein States and Dimerization of Pd-L1. J. Med. Chem. 2017, 60, 5857–5867. [Google Scholar] [CrossRef]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-Molecule Inhibitors of Pd-1/Pd-L1 Immune Checkpoint Alleviate The Pd-L1-Induced Exhaustion of T-Cells. Oncotarget 2017, 8, 72167–72181. [Google Scholar] [CrossRef]
- Perry, E.; Mills, J.J.; Zhao, B.; Wang, F.; Sun, Q.; Christov, P.P.; Tarr, J.C.; Rietz, T.A.; Olejniczak, E.T.; Lee, T.; et al. Fragment-Based Screening of Programmed Death Ligand 1 (Pd-L1). Bioorg. Med. Chem. Lett. 2019. [Google Scholar] [CrossRef]
- Zhan, M.M.; Hu, X.Q.; Liu, X.X.; Liu, X.X.; Ruan, B.F.; Xu, J.; Liao, C. From Monoclonal Antibodies to Small Molecules: The Development of Inhibitors Targeting the Pd-1/Pd-L1 Pathway. Drug Discov. Today 2016, 21, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Integrity. Available online: Https://Integrity.Clarivate.Com/ (accessed on 20 January 2019).
- Reaxys. Available online: Https://Integrity.Clarivate.Com/ (accessed on 20 January 2019).
- Villar, E.A.; Beglov, D.; Chennamadhavuni, S.; Porco, J.A., Jr.; Kozakov, D.; Vajda, S.; Whitty, A. How Proteins Bind Macrocycles. Nat. Chem. Biol. 2014, 10, 723–731. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |













| Pharmacophore | Number of Features | Feature Set | Selectivity Score | Sensitivity * | Specificity * |
|---|---|---|---|---|---|
| Pharmacophore01 | 5 | DHHHR | 8.8639 | 0.703 | 0.993 |
| Pharmacophore02 | 5 | ADHHR | 8.8639 | 0.703 | 0.919 |
| Pharmacophore03 | 5 | ADHHR | 8.8639 | 0.741 | 0.993 |
| Pharmacophore04 | 5 | ADHHR | 8.8639 | 0.667 | 0.980 |
| Pharmacophore05 | 5 | ADHHR | 8.8639 | 0.889 | 0.966 |
| Pharmacophore06 | 5 | ADHHR | 8.8639 | 0.741 | 0.993 |
| Pharmacophore07 | 5 | AHHHR | 7.9504 | 0.778 | 0.838 |
| Parameter | Values |
|---|---|
| The number of molecules in the database | 175 |
| The number of actives in the database | 27 |
| The number of hit molecules from the database | 21 |
| The number of active molecules in the hit list | 20 |
| False negatives | 6 |
| False positives | 1 |
| % Yield of actives | 74.1 |
| % Sensitivity | 74.1 |
| % Specificity | 99.3 |
| Compounds | BMS-1166 | BMS-1001 | BMS-202 | 28131141 | BMS-200 | 28131140 | BMS-242 | BMS-37 | 28131145 | 28131143 | R * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | 1.4 | 2.25 | 18 | 43 | 80 | 6-100 | 6–100 | 6–100 | 110–1000 | 110–1000 | |
| Model01 Fit Value | 3.81036 | 4.48433 | none | 3.71533 | 3.80462 | 2.80741 | none | none | 1.56344 | none | 0.655 |
| Model02 Fit Value | 4.16447 | 3.93019 | 2.68308 | 3.7521 | 2.98747 | 2.73383 | 2.6811 | 2.67567 | 3.04527 | 2.58668 | 0.819 |
| Model03 Fit Value | 4.42391 | 4.52196 | 3.53526 | 3.90221 | 3.99688 | 3.30513 | 3.6466 | 3.95454 | 4.30285 | 4.01664 | 0.565 |
| Model04 Fit Value | 4.14129 | 4.63203 | 1.71529 | 2.9739 | 4.17607 | 2.10849 | 2.35671 | 1.76556 | 4.33115 | 2.77859 | 0.47 |
| Model05 Fit Value | 4.1626 | 4.4831 | 3.1851 | 3.27722 | 3.38762 | 3.0444 | 3.51356 | 2.73352 | 3.67542 | 3.56603 | 0.781 |
| Model06 Fit Value | 4.12881 | 4.49658 | 1.69069 | 3.43773 | 3.9225 | 2.71941 | 1.94917 | 1.82323 | 3.79388 | 2.59189 | 0.529 |
| Model07 Fit Value | 4.5101 | 4.72431 | 2.82402 | 3.79522 | 2.07746 | 3.38077 | 3.46818 | 3.61272 | 3.98516 | 3.93575 | 0.539 |
| Model08 Fit Value | 4.47768 | 4.73352 | 3.58699 | 4.03414 | 3.35641 | 3.77544 | 4.08634 | 4.03562 | 3.35984 | 4.07816 | 0.679 |
| Model09 Fit Value | 3.82049 | 4.05292 | 0.09738 | 4.00387 | 4.45517 | 2.82319 | 1.18001 | 0.17939 | 2.31305 | 0.11979 | 0.428 |
| Model10 Fit Value | 3.30267 | 4.09233 | 1.80806 | 3.41994 | 4.28178 | 2.67542 | 2.27937 | 1.80307 | 3.69411 | 2.88655 | 0.28 |
| Parameter | Values |
|---|---|
| The number of molecules in the database | 260 |
| The number of actives in the database | 110 |
| The number of hit molecules from the database | 78 |
| The number of active molecules in the hit list | 78 |
| False negatives | 32 |
| False positives | 0 |
| % Yield of actives | 70.9 |
| % Sensitivity | 70.9 |
| % Specificity | 100 |
| Name | Structure | IC50 (nM) |
|---|---|---|
| BMS-1166 |  | 1.4 |
| BMS-1001 |  | 2.25 |
| BMS-202 |  | 18 |
| 28131141 |  | 43 |
| BMS-200 |  | 80 |
| 28131140 |  | 6-100 |
| BMS-242 |  | 6-100 |
| BMS-37 |  | 6-100 |
| 28131135 |  | 6-100 |
| 28131138 |  | 6-100 |
| BMS-8 |  | 146 |
| 28131145 |  | 110-1000 |
| 28131143 |  | 110-1000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Y.; Li, X.; Yao, H.; Lin, K. The Characteristics of PD-L1 Inhibitors, from Peptides to Small Molecules. Molecules 2019, 24, 1940. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24101940
Zhong Y, Li X, Yao H, Lin K. The Characteristics of PD-L1 Inhibitors, from Peptides to Small Molecules. Molecules. 2019; 24(10):1940. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24101940
Chicago/Turabian StyleZhong, Yanwen, Xuanyi Li, Hequan Yao, and Kejiang Lin. 2019. "The Characteristics of PD-L1 Inhibitors, from Peptides to Small Molecules" Molecules 24, no. 10: 1940. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24101940