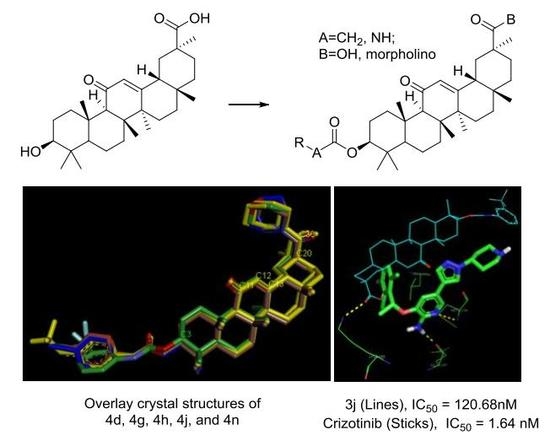
3.2. Chemistry
3.2.1. 3β-hydroxy-30-morpholino-olean-12-ene-11,30-dione 2
The 18β-GA (0.47 g, 1.0 mmol) was dissolved in acetonitrile (20 mL), then EDCI (0.23 g, 1.2 mmol), triethylamine (0.13 g, 1.2 mmol) and HOBt (0.16 g, 1.2 mmol) were added. The mixture was stirred under reflux for 20 min. Then morpholine (0.11 g, 1.2 mmol) was added, and the mixture was stirred under reflux for 24 h. The solvent was removed under reduced pressure to give a residue which was partitioned between C2H5OH and H2O. The solution was stirred at room temperature for 30 min, and a solid was obtained by filtration while washing with H2O.
A white solid; Yield, 93.9%; m.p. 239.6–240.8 °C; 1H-NMR (400 MHz, Chloroform-d) δ 5.67 (s, 1H, CH-12), 3.70–3.53 (m, 8H, morpholine-H), 3.20 (dd, J = 10.8, 5.4 Hz, 1H, OH-3), 2.77 (dt, J = 13.4, 3.6 Hz, 1H, CH-1), 2.31 (s, 1H, CH-9), 2.27 (dd, J = 13.5, 3.7 Hz, 1H, CH-16), 1.34 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.11 (s, 3H, CH3-26), 1.10 (s, 3H, CH3-29), 0.98 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.78 (s, 3H, CH3-28), 0.68 (d, J = 11.4 Hz, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.12 (C11), 174.01 (C30), 169.45 (C13), 128.56 (C12), 78.75 (C3), 66.93 (morpholine C), 61.78 (C9), 54.92 (C5), 48.15 (C18), 45.26 (C14), 43.79 (C20), 43.70 (morpholine C), 43.26 (C8/19), 39.13 (C1), 39.10 (C4), 37.68 (C22), 37.06 (C10), 33.22 (C7), 32.79 (C17), 31.76 (C21), 28.39 (C29), 28.07 (C28), 27.27 (C23), 26.96 (C2), 26.70 (C15), 26.41 (C16), 23.14 (C27), 18.66 (C26), 17.46 (C6), 16.37 (C25), 15.56 (C24); HRMS (m/z): [M + H]+ calcd. for C34H54NO4: 540.40528, found: 540.40440.
1H-benzo[d][1,2,3]triazol-1-yl-3β-hydroxy-11-oxo-olean-12-en-30-oate (
2a), white solid; Yield, 95.8%; m.p. 251.1–252.2 °C, (literature [
43]: 192–195 °C, decomp.);
1H-NMR (400 MHz, Chloroform-
d) δ 8.08 (d,
J = 8.4 Hz, 1H, phenyl), 7.56 (t,
J = 7.6 Hz, 1H, phenyl), 7.44 (t,
J = 7.7 Hz, 1H, phenyl), 7.34 (d,
J = 8.3 Hz, 1H, phenyl), 5.71 (s, 1H, CH-12), 3.22 (dd,
J = 10.7, 5.5 Hz, 1H, OH-3), 2.77 (dt,
J = 13.5, 3.6 Hz, 1H, CH-1), 2.39–2.23 (m, 2H, CH-9/16), 1.41 (s, 3H, CH
3-27), 1.15 (s, 3H, CH
3-25), 1.13 (s, 3H, CH
3-26), 1.00 (s, 3H, CH
3-29), 0.93 (s, 3H, CH
3-23), 0.80 (s, 3H, CH
3-24), 0.72 (d,
J = 11.6 Hz, 1H, CH-5);
13C-NMR (101 MHz, Chloroform-
d) δ 199.91 (C11), 172.51 (C30), 167.60 (C13), 143.54 (phenyl), 129.02 (phenyl), 128.83 (phenyl), 128.54 (C12), 124.82 (phenyl), 120.66, (phenyl) 107.81 (phenyl), 78.70 (C3), 61.83 (C9), 54.89 (C5), 48.20 (C18), 45.36 (C20), 44.36(C8), 43.15(C19), 40.85 (C1), 39.11 (C4), 37.75 (C22), 37.06 (C10), 32.72 (C7), 31.97 (C17), 31.16 (C21), 28.54 (C29), 28.08 (C28), 28.02 (C23), 27.25 (C2), 26.34 (C15/16), 23.48 (C27), 18.67 (C26), 17.45 (C6), 16.34 (C25), 15.57 (C24); HRMS (
m/
z): [M + H]
+ calcd. for C
36H
50N
3O
4: 588.38013, found: 588.38018.
3.2.2. General Procedure for Preparation of Carbamate Derivatives (3a–3o)
A mixture of 18β-GA 1 (0.19 g, 0.40 mmol) and substituted isocyanates (0.48 mmol) in ethyl acetate was stirred under reflux for 24 h. The organic layer was washed with 10% aqueous hydrochloric acid, 5% of aqueous NaHCO3, brine and was dried over anhydrous Na2SO4. The organic layer was then concentrated under reduced pressure. The residue obtained was purified by silica gel column chromatography using CH2Cl2–CH3OH as the eluent.
3β-(((3,4-dichlorophenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3a), white solid; Yield, 88.5%; m.p. 260.0–261.4 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.63 (s, 1H, phenyl-H), 7.33 (d, J = 8.7 Hz, 1H, phenyl-H), 7.18 (d, J = 8.8 Hz, 1H, phenyl-H), 6.71 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.50 (dd, J = 10.5, 5.9 Hz, 1H, CH-3), 2.87–2.77 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.18 (dd, J = 13.8, 4.1 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28),0.80 (d, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.33 (C11), 181.32 (C30), 169.61 (C13), 137.62 (phenyl), 132.79 (phenyl), 130.45 (phenyl), 128.38 (C12), 126.31 (phenyl), 120.80 (phenyl), 117.72 (phenyl), 88.24 (C3), 61.62 (C9), 55.04 (C5), 48.23 (C18), 45.44 (C14), 43.77 (C20), 43.20 (C8), 40.81 (C19), 38.70 (C1), 38.23 (C4), 37.67 (C22), 36.88 (C10), 32.64 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.09 (C23), 26.44 (C2), 26.36 (C15), 23.85 (C16), 23.36 (C27), 18.65 (C26), 17.33 (C6), 16.81 (C25), 16.39 (C24); HRMS (m/z): [M + H]+ calcd. for C37H50Cl2NO5: 658.30660, found: 658.31669.
3β-(((4-chloro-3-(trifluoromethyl)phenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3b), white solid; Yield, 87.0%; m.p. 247.8–248.5 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.76 (s, 1H, phenyl-H), 7.54 (s, 1H, phenyl-H), 7.40 (d, J = 8.7 Hz, 1H, phenyl-H), 6.83 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.56–4.47 (m, 1H, CH-3), 2.82 (d, J = 13.6 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.17 (d, J = 11.9 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28),0.80 (d, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.30 (C11), 181.19 (C30), 169.61 (C13), 137.02 (phenyl), 131.94 (phenyl), 128.90 (phenyl), 128.58 (phenyl), 128.38 (C12), 125.71(CF3), 123.90 (CF3), 82.55 (C9), 61.63 (C9), 55.05 (C5), 48.23 (C18), 45.44 (C14), 43.76 (C20), 43.19 (C8), 40.81 (C19), 38.70 (C1), 38.22 (C4), 37.67 (C22), 36.87 (C10), 32.63 (C7), 31.85 (C17), 30.89 (C21), 28.52 (C29), 28.42 (C28), 28.08 (C23), 26.44 (C2), 26.36 (C15), 23.84 (C16), 23.35 (C27), 18.65 (C26), 17.33 (C6), 16.78 (C25), 16.39 (C24); HRMS (m/z): [M + H]+ calcd. for C38H50ClF3NO5: 692.33296, found: 692.33792.
3β-(((3,5-dichlorophenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3c), white solid; Yield, 90.5%; m.p. 236.1–237.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.35 (s, 2H, phenyl-H), 7.02 (t, J = 1.8 Hz, 1H, phenyl-H), 6.62 (s, 1H, N-H), 5.69 (s, 1H, CH-12), 4.50 (dd, J = 10.7, 5.7 Hz, 1H, CH-3), 2.82 (d, J = 13.2 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.17 (d, J = 11.9 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.79–0.73 (m, 1H, CH-5); 13C-NMR (101 MHz, DMSO-d6) δ199.36 (C11), 178.12 (C30), 170.33 (C13), 153.76 (carbamoyl), 142.30 (phenyl), 134.55 (phenyl), 127.62 (C12), 121.83 (phenyl), 116.62 (phenyl), 81.34 (C3), 61.24 (C9), 56.24 (C5), 48.48 (C18), 45.29 (C14), 43.50 (C20), 43.39 (C8), 41.03 (C19), 38.32 (C1/4), 37.94 (C22), 36.91 (C10), 32.56 (C7), 31.96 (C17), 30.02 (C21), 28.82 (C29), 28.24 (C28), 28.09 (C23), 26.51 (C2), 26.20 (C15), 23.91 (C16), 23.44 (C27), 18.73 (C26), 17.34 (C6), 17.15 (C25), 16.65 (C24); HRMS (m/z): [M + H]+ calcd. for C37H50Cl2NO5: 658.30660, found: 658.31238.
3β-(((4-chlorophenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3d), white solid; Yield, 87.3%; m.p. 235.1–236.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.33 (d, J = 8.4 Hz, 2H, phenyl-H), 7.25 (d, J = 2.6 Hz, 2H, phenyl-H), 6.66 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.58–4.43 (m, 1H, CH-3), 2.81 (dt, J = 13.5, 3.6 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.23–2.13 (m, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.35 (C11), 181.38 (C30), 169.57 (C13), 136.66 (phenyl), 128.97 (phenyl), 128.38 (C12), 128.15 (phenyl), 119.68 (phenyl), 81.95 (C3), 61.64 (C9), 55.04 (C5), 48.22 (C18), 45.44 (C14), 43.77 (C20), 43.19 (C8), 40.81 (C19), 38.72 (C1), 38.24 (C4), 37.68 (C22), 36.88 (C10), 32.65 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.08 (C23), 26.44 (C2), 26.36 (C15), 23.88 (C16), 23.35 (C27), 18.65 (C26), 17.34 (C6), 16.82 (C25), 16.40 (C24); HRMS (m/z): [M + H]+ calcd. for C37H51ClNO5: 624.34558, found: 624.35065.
3β-(((3-chlorophenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3e), white solid; Yield, 89.9%; m.p. 268.0.0–269.5 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.51 (s, 1H, phenyl-H), 7.20 (d, J = 4.9 Hz, 2H, phenyl-H), 7.00 (td, J = 4.5, 2.0 Hz, 1H, phenyl-H), 6.69 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.50 (dd, J = 10.4, 6.1 Hz, 1H, CH-3), 2.82 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.18 (dd, J = 13.0, 3.7 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.82-0.81 (m, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.34 (C11), 181.33 (C30), 169.56 (C13), 153.35 (carbamoyl), 139.25 (phenyl), 134.70 (phenyl), 129.94 (phenyl), 128.39 (C12), 123.22 (phenyl), 118.52 (phenyl),116.46 (phenyl), 82.11 (C3), 61.64 (C9), 55.04 (C5), 48.22 (C18), 45.44 (C14), 43.77 (C20), 43.19 (C8), 40.81 (C19), 38.72 (C1), 38.24 (C4), 37.68 (C22), 36.88 (C10), 32.65 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.08 (C23), 26.44 (C2), 26.36 (C15), 23.86 (C16), 23.36 (C27), 18.65 (C26), 17.34 (C6), 16.82 (C25), 16.39 (C24); HRMS (m/z): [M + H]+ calcd. for C37H51ClNO5: 624.34558, found: 624.34960.
3β-(((3-chloro-4-methylphenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3f), white solid; Yield, 90.4%; m.p. 270.7–271.6 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.48 (s, 1H, phenyl-H), 7.11 (s, 2H, phenyl-H), 6.60 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.49 (t, J = 8.3 Hz, 1H, CH-3), 2.81 (dt, J = 13.4, 3.6 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.29 (s, 3H, phenyl-CH3), 2.18 (dd, J = 13.3, 4.2 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (m, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.36 (C11), 181.39 (C30), 169.56 (C13), 153.48 (carbamoyl), 136.83 (phenyl), 134.49 (phenyl), 130.95 (phenyl), 130.62 (phenyl), 128.39 (C12), 119.22 (phenyl), 116.83 (phenyl), 81.85 (C3), 61.64 (C9), 55.05 (C5), 48.22 (C18), 45.45 (C14), 43.77 (C20), 43.19 (C8), 40.80 (C19), 38.73 (C1), 38.24 (C4), 37.68 (C22), 36.89 (C10), 32.65 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.08 (C23), 26.44 (C2), 26.37 (C15), 23.87 (C16), 23.36 (C27), 19.33 (phenyl-CH3), 18.65 (C26), 17.34 (C6), 16.82 (C25), 16.39 (C24); HRMS (m/z): [M + Na]+ calcd. for C38H52ClNNaO5: 660.34317, found: 660.34747.
3β-(((4-bromophenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3g), white solid; Yield, 87.2%; m.p. 263.0–265.0 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.42–7.35 (m, 2H, phenyl-H), 7.27 (d, J = 8.4 Hz, 2H, phenyl-H), 6.65 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.50 (t, J = 8.3 Hz, 1H, CH-3), 2.85–2.77 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.18 (dd, J = 13.7, 4.1 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.34 (C11), 181.30 (C30), 169.56 (C13), 153.36 (carbamoyl), 137.17 (phenyl), 131.91 (phenyl), 128.39 (C12), 120.05 (phenyl), 81.92 (C3), 61.64 (C9), 55.04 (C5), 48.22 (C18), 45.44 (C14), 43.77 (C20), 43.19 (C8), 40.81 (C19), 38.72 (C1), 38.24 (C4), 37.68 (C22), 36.88 (C10), 32.65 (C7), 31.85 (C17), 30.89 (C21), 28.52 (C29), 28.43 (C28), 28.08 (C23), 26.44 (C2), 26.36 (C15), 23.87 (C16), 23.35 (C27), 18.65 (C26), 17.34 (C6), 16.82 (C25), 16.40 (C24); HRMS (m/z): [M + H]+ calcd. for C37H51BrNO5: 668.29506, found: 668.30334, 670.30211.
3β-(((4-fluorophenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3h), white solid; Yield, 88.6%; m.p. 267.9–268.9 °C; 1H-NMR (400 MHz, DMSO-d6) δ 12.25 (s, 1H, COOH), 9.57 (s, 1H, N-H), 7.51 (s, 2H, phenyl-H), 7.14 (t, J = 8.9 Hz, 2H, phenyl-H), 5.44 (s, 1H, CH-12), 4.42 (dd, J = 11.8, 4.6 Hz, 1H, CH-3), 2.73–2.65 (m, 1H, CH-1), 2.46 (s, 1H, CH-9), 2.18–2.04 (m, 2H, CH-16, CH-2), 1.41 (s, 3H, CH3-27), 1.13 (s, 3H, CH3-25), 1.11 (s, 3H, CH3-26), 1.08 (s, 3H, CH3-29), 0.92 (s, 3H, CH3-23), 0.91 (s, 3H, CH3-24), 0.79 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 200.36 (C11), 181.39 (C30), 169.55 (C13), 153.81 (carbamoyl), 134.01 (phenyl), 128.39 (C12), 120.13 (phenyl), 115.59 (d, J = 22 Hz, phenyl), 81.63 (C3), 61.65 (C9), 55.04 (C5), 48.22 (C18), 45.45 (C14), 43.77 (C20), 43.19 (C8), 40.81 (C19), 38.73 (C1), 38.25 (C4), 37.68 (C22), 36.88 (C10), 32.66 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.07 (C23), 26.44 (C2), 26.36 (C15), 23.89 (C16), 23.35 (C27), 18.65 (C26), 17.34 (C6), 16.81 (C25), 16.40 (C24); HRMS (m/z): [M + H]+ calcd. for C37H51FNO5: 608.37513, found: 608.38190.
3β-(((4-(trifluoromethyl)phenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3i), white solid; Yield, 90.8%; m.p. 242.1–242.9 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.52 (q, J = 8.7 Hz, 4H, phenyl-H), 6.87 (s, 1H, N-H), 5.71 (s, 1H, CH-12), 4.52 (dd, J = 10.5, 5.8 Hz, 1H, CH-3), 2.82 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.18 (dd, J = 13.5, 4.1 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.95 (s, 3H, CH3-23), 0.89 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.81 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.35 (C11), 181.40 (C30), 169.64 (C13), 153.37 (carbamoyl), 141.21 (phenyl), 128.37 (C12), 126.29 (q, J = 3.8 Hz, CF3), 126.27 (CF3), 126.23 (CF3), 125.50 (phenyl), 125.13 (phenyl), 124.80 (phenyl), 122.80 (phenyl), 117.97 (phenyl), 82.26 (C3), 61.64 (C9), 55.05 (C5), 48.23 (C18), 45.45 (C14), 43.78 (C20), 43.20 (C8), 40.81 (C19), 38.72 (C1), 38.24 (C4), 37.68 (C22), 36.88 (C10), 32.64 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.08 (C23), 26.44 (C2), 26.36 (C15), 23.86 (C16), 23.35 (C27), 18.65 (C26), 17.33 (C6), 16.83 (C25), 16.40 (C24); HRMS (m/z): [M + H]+ calcd. for C38H51F3NO5: 658.37193, found: 658.37843.
3β-(((3-(trifluoromethyl)phenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3j), white solid; Yield, 89.2%; m.p. 239.1–240.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.72 (s, 1H, phenyl-H), 7.40 (t, J = 8.0 Hz, 1H, phenyl-H), 7.28 (d, J = 7.8 Hz, 1H, phenyl-H), 6.82 (s, 1H, N-H), 5.71 (s, 1H, CH-12), 4.52 (t, J = 8.3 Hz, 1H, CH-3), 2.86–2.78 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.23–2.13 (m, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.95 (s, 3H, CH3-23), 0.89 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.81 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.32 (C11), 181.25 (C30), 169.57 (C13), 153.42 (carbamoyl), 138.63 (phenyl), 131.56 (CF3), 131.24 (CF3), 130.91 (CF3), 129.50 (phenyl), 128.39 (C12), 125.21 (phenyl), 122.50 (phenyl), 119.76 (phenyl), 82.36 (C3), 61.64 (C9), 55.06 (C5), 48.23 (C18), 45.44 (C14), 43.77 (C20), 43.19 (C8), 40.81 (C19), 38.72 (C1), 38.22 (C4), 37.68 (C22), 36.88 (C10), 32.64 (C7), 31.85 (C17), 30.89 (C21), 28.52 (C29), 28.43 (C28), 28.09 (C23), 26.44 (C2), 26.36 (C15), 23.85 (C16), 23.36 (C27), 18.65 (C26), 17.34 (C6), 16.79 (C25), 16.39 (C24); HRMS (m/z): [M + H]+ calcd. for C38H51F3NO5: 658.37193, found: 658.37843.
3β-(((3,5-bis (trifluoromethyl)phenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3k), white solid; Yield, 87.0%; m.p. 248.8–251.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.90 (s, 2H, phenyl-H), 7.53 (s, 1H, phenyl-H), 6.97 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.53 (dd, J = 10.6, 5.9 Hz, 1H, CH-3), 2.84 (dd, J = 10.2, 3.4 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.22–2.13 (m, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.13 (s, 3H, CH3-29), 0.95 (s, 3H, CH3-23), 0.89 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.81 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.21 (C11), 180.40 (C30), 169.51 (C13), 153.11 (carbamoyl), 139.60 (phenyl), 132.52 (phenyl), 132.19 (phenyl), 128.41 (C12), 124.43 (CF3), 121.72 (CF3), 118.07 (phenyl), 116.43 (phenyl), 82.59 (C3), 61.61 (C9), 55.06 (C5), 48.23 (C18), 45.42 (C14), 43.72 (C20), 43.20 (C8), 40.85 (C19), 38.69 (C1), 38.20 (C4), 37.66 (C22), 36.87 (C10), 32.63 (C7), 31.85 (C17), 30.92 (C21), 28.51 (C29), 28.39 (C28), 28.10 (C23), 26.44 (C2), 26.36 (C15), 23.81(C16), 23.36 (C27), 18.65 (C26), 17.33 (C6), 16.77(C25), 16.38 (C24); HRMS (m/z): [M + H]+ calcd. for C39H50F6NO5: 726.35932, found: 726.36406.
3β-(((3-methoxyphenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3l), white solid; Yield, 86.9%; m.p. 259.5–261.1°C; 1H-NMR (400 MHz, Chloroform-d) δ 7.17 (t, J = 8.2 Hz, 1H, phenyl-H), 6.84 (d, J = 8.0 Hz, 1H, phenyl-H), 6.65 (s, 1H, N-H), 6.59 (dd, J = 8.4, 2.4 Hz, 1H, phenyl-H), 5.70 (s, 1H, CH-12), 4.55–4.45 (m, 1H, CH-3), 3.79 (s, 3H, CH3), 2.85–2.76 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.18 (dd, J = 13.4, 4.1 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.37 (C11), 181.35 (C30), 169.53 (C13), 160.22 (phenyl), 153.37 (carbamoyl), 139.31 (phenyl), 129.67(phenyl), 128.39 (C12), 109.15 (phenyl), 81.72 (C3), 61.66 (C9), 55.25 (-OCH3), 55.08 (C5), 48.23 (C18), 45.45 (C14), 43.77 (C20), 43.18 (C8), 40.81 (C19), 38.76 (C1), 38.25 (C4), 37.68 (C22), 36.89 (C10), 32.67 (C7), 31.85 (C17), 30.88 (C21), 28.53 (C29), 28.44 (C28), 28.07 (C23), 26.44 (C2), 26.37 (C15), 23.89 (C16), 23.34 (C27), 18.66 (C26), 17.34 (C6), 16.83 (C25), 16.40 (C24); HRMS (m/z): [M + Na]+ calcd. for C38H53NNaO6: 642.37706, found: 642.37890.
3β-(((4-methoxyphenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3m), white solid; Yield, 89.3%; m.p. 264.0–264.8 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.28 (s, 2H, phenyl-H), 6.83 (d, J = 8.9 Hz, 2H, phenyl-H), 6.50 (s, 1H, N-H), 5.70 (s, 1H, CH-12),4.48 (t, J = 8.2 Hz, 1H, CH-3), 3.76 (s, 3H, CH3), 2.80 (dt, J = 13.5, 3.7 Hz, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.22–2.13 (m, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.86 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.39 (C11), 181.40 (C30), 169.53 (C13), 153.85 (carbamoyl), 131.13 (phenyl), 128.39 (C12), 120.33 (phenyl), 114.16 (phenyl), 81.45 (C3), 61.66 (C9), 55.48 (-OCH3), 55.04 (C5), 48.21 (C18), 45.45 (C14), 43.77 (C20), 43.18 (C8), 40.81 (C19), 38.74 (C1), 38.26 (C4), 37.69 (C22), 36.89 (C10), 32.67 (C7), 31.85 (C17), 30.89 (C21), 28.52 (C29), 28.44 (C28), 28.07 (C23), 26.44 (C2), 26.37 (C15), 23.91 (C16), 23.35 (C27), 18.65 (C26), 17.34 (C6), 16.81 (C25), 16.39 (C24); HRMS (m/z): [M + Na]+ calcd. for C38H53NNaO6: 642.37706, found: 642.38301.
3β-(((4-(trifluoromethoxy)phenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3n), white solid; Yield, 92.1%; m.p. 256.4–258.4°C; 1H-NMR (400 MHz, Chloroform-d) δ 7.40 (d, J = 8.5 Hz, 2H, phenyl-H), 7.14 (d, J = 8.6 Hz, 2H, phenyl-H), 6.72 (s, 1H, N-H), 5.70 (s, 1H, CH-12), 4.58–4.42 (m, 1H, CH-3), 2.85–2.76 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.18 (dd, J = 13.5, 4.1 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.36 (C11), 181.49 (C30), 169.62 (C13), 153.29 (carbamoyl), 144.53(phenyl), 136.79 (phenyl), 128.37 (C12), 124.28 (CF3), 121.84 (phenyl), 121.73 (phenyl), 119.47 (phenyl), 119.18 (phenyl), 81.99 (C3), 61.64 (C9), 55.04 (C5), 48.22 (C18), 45.44 (C14), 43.78 (C20), 43.19 (C8), 40.80 (C19), 38.72 (C1), 38.25 (C4), 37.68 (C22), 36.88 (C10), 32.65 (C7), 31.85 (C17), 30.88 (C21), 28.52 (C29), 28.44 (C28), 28.06 (C23), 26.44 (C2), 26.36 (C15), 23.87 (C16), 23.35 (C27), 18.65 (C26), 17.33 (C6), 16.81(C25), 16.39 (C24); HRMS (m/z): [M + H]+ calcd. for C38H51F3NO6: 674.36685, found: 674.37311.
3β-(((3,5-dimethylphenyl)carbamoyl)oxy)-11-oxo-olean-12-en-30-oic acid (3o), white solid; Yield, 92.1%; m.p. 279.8–280.6 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.01 (s, 2H, phenyl-H), 6.68 (s, 1H, phenyl-H), 6.55 (s, 1H, N-H), 5.71 (s, 1H, CH-12), 4.49 (t, J = 8.2 Hz, 1H, CH-3), 2.85–2.76 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.27 (s, 6H, phenyl-CH3), 2.18 (dd, J = 13.5, 4.1 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.38 (C11), 181.43 (C30), 169.53 (C13), 153.61 (carbamoyl), 138.68 (phenyl), 137.84 (phenyl), 128.40 (C12), 124.95 (phenyl), 119.26 (phenyl), 81.69 (C3), 61.65 (C9), 55.07 (C5), 48.22 (C18), 45.45 (C14), 43.77 (C20), 43.19 (C8), 40.81 (C19), 38.75 (C1), 38.24 (C4), 37.69 (C22), 36.90 (C10), 32.67 (C7), 31.85 (C17), 30.89 (C21), 28.53 (C29), 28.44 (C28), 28.08 (C23), 26.44 (C2), 26.37 (C15), 23.90 (C16), 23.37 (C27), 21.37 (phenyl-CH3), 21.34 (phenyl-CH3), 18.66 (C26), 17.35 (C6), 16.82 (C25), 16.39 (C24); HRMS (m/z): [M + Na]+ calcd. for C39H55NNaO5: 640.39779, found: 640.340185.
3.2.3. General Procedure for Preparation of Carbamate Derivatives (4a–4o)
A mixture of compound 2 (0.22 g, 0.40 mmol) and substituted isocyanates (0.48 mmol) in ethyl acetate was stirred under reflux for 24 h. The organic layer was washed with 10% aqueous hydrochloric acid, 5% of aqueous NaHCO3, brine and was dried over anhydrous Na2SO4. The organic layer was then concentrated under reduced pressure. The residue obtained was purified by silica gel column chromatography using CH2Cl2–CH3OH as the eluent.
3β-(((3,4-dichlorophenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4a), white solid; Yield, 93.0%; m.p.292.5–293.0 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.63 (s, 1H, phenyl-H), 7.32 (d, J = 8.8 Hz, 1H, phenyl-H), 7.18 (d, J = 8.8 Hz, 1H, phenyl-H), 6.72 (s, 1H, N-H), 5.68 (s, 1H, CH-12), 4.48 (dd, J = 10.8, 5.7 Hz, 1H, CH-3), 3.65 (t, J = 6.3 Hz, 8H, morpholine-H), 2.81 (dt, J = 13.4, 3.8 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.27 (d, J = 12.7 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.10 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.86 (s, 3H, CH3-24), 0.80 (s, 3H, CH3-28), 0.79 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 199.99 (C11), 174.02 (C30), 169.71 (C13), 153.23 (carbamoyl), 137.66 (phenyl), 132.78 (phenyl), 130.44 (phenyl), 128.46 (C12), 126.27 (phenyl), 120.13 (phenyl), 117.70 (phenyl), 82.25 (C3), 66.93 (morpholine C), 61.64 (C9), 55.05 (C5), 48.20 (C18), 45.28 (C14), 43.79 (C20), 43.66 (morpholine C), 43.29 (C8), 38.74 (C1), 38.22 (C4), 37.67 (C22), 36.89 (C10), 33.23 (morpholine C), 32.69 (C7), 31.77 (C17), 28.41 (C28), 28.08 (C23), 26.96 (C2), 26.68 (C15), 26.40 (C16), 23.84 (C29), 23.11(C27), 18.66 (C26), 17.34 (C6), 16.79 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C41H56Cl2N2NaO5: 749.34640, found: 749.34901.
3β-(((4-chloro-3-(trifluoromethyl)phenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4b), white solid; Yield, 92.5%; m.p. 209.5–211.1 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.77 (d, J = 2.5 Hz, 1H, phenyl-H), 7.54 (d, J = 8.6 Hz, 1H, phenyl-H), 7.40 (d, J = 8.7 Hz, 1H, phenyl-H), 6.85 (s, 1H, N-H), 5.68 (s, 1H, CH-12), 4.50 (dd, J = 11.0, 5.5 Hz, 1H, CH-3), 3.70–3.56 (m, 8H, morpholine-H), 2.81 (dt, J = 13.7, 3.7 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.32–2.23 (m, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 199.97 (C11), 174.04 (C30), 169.70 (C13), 153.30 (carbamoyl), 137.09 (phenyl), 131.94 (phenyl), 128.90 (phenyl), 128.58 (phenyl), 128.48 (C12), 125.29 (q, J = 271 Hz, CF3), 121.21 (phenyl), 117.45 (phenyl), 82.43 (C3), 66.95 (morpholine C), 61.65 (C9), 55.08 (C5), 48.21 (C18), 45.29 (C14), 43.80 (C20), 43.68 (morpholine C), 43.30 (C8), 38.75 (C1), 38.22 (C4), 37.69 (C22), 36.90 (C10), 33.24 (morpholine C), 32.71 (C7), 31.78 (C17), 28.42 (C28), 28.10 (C23), 26.97 (C2), 26.70 (C15), 26.41 (C16), 23.85 (C29), 23.11(C27), 18.67(C26), 17.35 (C6), 16.79 (C25), 16.42 (C24); HRMS (m/z): [M + Na]+ calcd. for C42H56ClF3N2NaO5: 783.37275, found: 783.37677.
3β-(((3,5-dichlorophenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4c), white solid; Yield, 91.7%; m.p. 278.2–280.3 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.35 (s, 2H, phenyl-H), 7.01 (q, J = 2.7, 1.9 Hz, 1H, phenyl-H), 6.78 (s, 1H, N-H), 5.68 (s, 1H, CH-12), 4.48 (dd, J = 10.9, 5.5 Hz, 1H, CH-3), 3.65 (q, J = 6.4, 5.8 Hz, 8H, morpholine-H), 2.81 (dt, J = 13.4, 3.4 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.31–2.22 (m, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82(s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.97 (C11), 174.03 (C30), 169.69 (C13), 153.08 (carbamoyl), 140.05 (phenyl), 135.23 (phenyl), 128.47 (C12), 123.06 (phenyl), 116.67 (phenyl), 82.38 (C3), 66.94 (morpholine C), 61.64 (C9), 55.05 (C5), 48.21 (C18), 45.28 (C14), 43.79 (C20), 43.65 (morpholine C), 43.29 (C8), 38.73 (C1), 38.21 (C4), 37.67 (C22), 36.89 (C10), 33.25 (morpholine C), 32.69 (C7), 31.77 (C17), 28.41 (C28), 28.08 (C23), 26.96(C2), 26.68 (C15), 26.40 (C16), 23.82 (C29), 23.12 (C27), 18.66 (C26), 17.34 (C6), 16.79 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C41H56Cl2N2NaO5: 749.34640, found: 749.34951.
3β-(((4-chlorophenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4d), white solid; Yield, 92.8%; m.p. 298.4–299.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.33 (d, J = 8.6 Hz, 2H, phenyl-H), 7.24 (d, J = 7.8 Hz, 2H, phenyl-H), 6.64 (s, 1H, N-H), 5.68 (s, 1H, CH-12), 4.48 (dd, J = 10.6, 5.9 Hz, 1H, CH-3), 3.67-3.56 (m, 8H, morpholine-H), 2.80 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.31–2.22 (m, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82(s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.98 (C11), 174.01 (C30), 169.61 (C13), 153.46 (carbamoyl), 136.69 (phenyl), 128.97 (phenyl), 128.48 (C12), 128.13 (phenyl), 119.68 (phenyl), 81.88 (C3), 66.94 (morpholine C), 61.66 (C9), 55.06 (C5), 48.21 (C18), 45.28 (C14), 43.78 (C20), 43.64 (morpholine C), 43.28 (C8), 38.76 (C1), 38.23 (C4), 37.67 (C22), 36.89 (C10), 33.27 (morpholine C), 32.71 (C7), 31.77 (C17), 28.41 (C28), 28.07 (C23), 26.96 (C2), 26.69 (C15), 26.40 (C16), 23.87 (C29), 23.11 (C27), 18.66 (C26), 17.35 (C6), 16.80 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C41H57ClN2NaO5: 715.38537, found: 715.38855.
3β-(((3-chlorophenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4e), white solid; Yield, 94.4%; m.p. 291.0–292.5 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.52 (s, 1H, phenyl-H), 7.19 (d, J = 5.1 Hz, 2H, phenyl-H), 7.00 (dq, J = 7.1, 2.0 Hz, 1H, phenyl-H), 6.70 (s, 1H, N-H), 5.68 (s, 1H, CH-12), 4.49 (dd, J = 10.7, 5.9 Hz, 1H, CH-3), 3.69–3.56 (m, 8H, morpholine-H), 2.80 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.27 (d, J = 13.2 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82(s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.98 (C11), 174.02 (C30), 169.63 (C13), 153.34 (carbamoyl), 139.30 (phenyl), 134.70 (phenyl), 129.94 (phenyl), 128.48 (C12), 123.18 (phenyl), 118.53 (phenyl), 116.42 (phenyl), 80.20 (C3), 66.94 (morpholine C), 61.65 (C9), 55.06 (C5), 48.22 (C18), 45.28 (C14), 43.78 (C20), 43.63 (morpholine C), 43.28 (C8), 38.75 (C1), 38.22 (C4), 37.67 (C22), 36.89 (C10), 33.28 (morpholine C), 32.70 (C7), 31.77 (C17), 28.41 (C28), 28.07 (C23), 26.96 (C2), 26.69 (C15), 26.40 (C16), 23.86 (C29), 23.11 (C27), 18.66 (C26), 17.35 (C6), 16.80 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C41H57ClN2NaO5: 715.38537, found: 715.38920.
3β-(((3-chloro-4-methylphenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4f), white solid; Yield, 91.8%; m.p. 293.7–295.6 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.50 (s, 1H, phenyl-H), 7.12 (s, 2H, phenyl-H), 6.57 (s, 1H, N-H), 5.69 (s, 1H, CH-12), 4.49 (dd, J = 10.3, 6.1 Hz, 1H, CH-3), 3.64 (qd, J = 8.5, 8.1, 3.5 Hz, 8H, morpholine-H), 2.81 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.36 (s, 1H, CH-9), 2.33–2.28 (m, 3H, CH3), 2.25 (d, J = 3.2 Hz, 1H, CH-16), 1.36 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.83 (s, 1H, CH-5), 0.81 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.98 (C11), 174.02 (C30), 169.59 (C13), 153.45 (carbamoyl), 136.85 (phenyl), 134.50 (phenyl), 130.95 (phenyl), 130.62 (phenyl), 128.49 (C12), 119.10 (phenyl), 116.78 (phenyl), 81.85 (C3), 66.95 (morpholine C), 61.67 (C9), 55.07 (C5), 48.23 (C18), 45.29 (C14), 43.78 (C20), 43.62 (morpholine C), 43.28 (C8), 38.77 (C1), 38.23 (C4), 37.67 (C22), 36.90 (C10), 33.30 (morpholine C), 32.71 (C7), 31.77 (C17), 28.41 (C28), 28.08 (C23), 26.97 (C2), 26.69 (C15), 26.41 (C16), 23.87 (C29), 23.12 (C27), 19.32 (CH3), 18.67 (C26), 17.35 (C6), 16.80 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C42H59ClN2NaO5: 729.40102, found: 729.40510.
3β-(((4-bromophenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4g), white solid; Yield, 90.6%; m.p. 309.7–3101 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.38 (dd, J = 8.9, 2.1 Hz, 2H, phenyl-H), 7.28 (d, J = 8.4 Hz, 2H, phenyl-H), 6.66 (s, 1H, N-H), 5.68 (d, J = 1.9 Hz, 1H, CH-12), 4.48 (dd, J = 11.3, 5.5 Hz, 1H, CH-3), 3.73–3.55 (m, 8H, morpholine-H), 2.85–2.75 (m, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.27 (d, J = 12.8 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.10 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.82(s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.97 (C11), 174.01 (C30), 169.62 (C13), 153.42 (carbamoyl), 137.22 (phenyl), 131.90 (phenyl), 128.48 (C12), 120.05 (phenyl), 115.62 (phenyl), 81.95 (C3), 66.94 (morpholine C), 61.66 (C9), 55.06 (C5), 48.21 (C18), 45.28 (C14), 43.78 (C20), 43.64 (morpholine C), 43.28 (C8), 38.76 (C1), 38.23 (C4), 37.67 (C22), 36.89 (C10), 33.26 (morpholine C), 32.70 (C7), 31.77 (C17), 28.41 (C28), 28.08 (C23), 26.96 (C2), 26.69 (C15), 26.40 (C16), 23.87 (C29), 23.11 (C27), 18.66 (C26), 17.35 (C6), 16.80 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C41H57BrN2NaO5: 759.33486, found: 759.33486, 761.33783.
3β-(((4-fluorophenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4h), white solid; Yield, 94.0%; m.p. 305.1–311.4 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.33 (s, 2H, phenyl-H), 6.97 (td, J = 8.6, 1.5 Hz, 2H, phenyl-H), 6.62 (s, 1H, N-H), 5.67 (s, 1H, CH-12), 4.48 (dd, J = 10.3, 6.1 Hz, 1H, CH-3), 3.65-3.59 (m, 8H, morpholine-H), 2.80 (dt, J = 14.1, 3.6 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.31–2.22 (m, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.86 (s, 3H, CH3-24), 0.82 (s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.99 (C11), 174.02 (C30), 169.60 (C13), 153.76 (carbamoyl), 134.05 (phenyl), 128.48 (C12), 120.14 (phenyl), 115.58 (d, J = 22.0 Hz, phenyl), 81.70 (C3), 66.94 (morpholine C), 61.67 (C9), 55.06 (C5), 48.21 (C18), 45.28 (C14), 43.78 (C20), 43.63 (morpholine C), 43.28 (C8), 38.77 (C1), 38.24 (C4), 37.67 (C22), 36.90 (C10), 33.27 (morpholine C), 32.71 (C7), 31.77 (C17), 28.41 (C28), 28.07 (C23), 26.96 (C2), 26.69 (C15), 26.40 (C16), 23.89 (C29), 23.10 (C27), 18.66 (C26), 17.35 (C6), 16.79 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C41H58FN2O5: 677.43298, found: 677.43850.
3β-(((4-(trifluoromethyl)phenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4i), white solid; Yield, 91.4%; m.p. 296.4–298.0 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.52 (q, J = 8.8 Hz, 4H, phenyl-H), 6.83 (s, 1H, N-H), 5.68 (s, 1H, CH-12), 4.51 (dd, J = 10.8, 5.6 Hz, 1H, CH-3), 3.70–3.56 (m, 8H, morpholine-H), 2.82 (dt, J = 13.6, 3.6 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.28 (dd, J = 13.9, 3.6 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.83 (s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.96 (C11), 174.01 (C30), 169.65 (C13), 153.23 (carbamoyl), 141.24 (phenyl), 128.48 (C12), 126.28 (q, J = 3.8 Hz, CF3), 125.50 (phenyl), 125.10 (phenyl), 124.78 (phenyl), 122.80 (phenyl), 117.93 (phenyl), 82.20 (C3), 66.94 (morpholine C), 61.65 (C9), 55.06 (C5), 48.20 (C18), 45.28 (C14), 43.78 (C20), 43.66 (morpholine C), 43.28 (C8), 38.75 (C1), 38.23 (C4), 37.67 (C22), 36.89 (C10), 33.24 (morpholine C), 32.70 (C7), 31.77 (C17), 28.41(C28), 28.08 (C23), 26.96 (C2), 26.68 (C15), 26.40 (C16), 23.85 (C29), 23.10 (C27), 18.66 (C26), 17.35 (C6), 16.80 (C25), 16.42 (C24); HRMS (m/z): [M + Na]+ calcd. for C42H57F3N2NaO5: 749.41173, found: 749.41688.
3β-(((3-(trifluoromethyl)phenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4j), white solid; Yield, 91.5%; m.p. 295.0–296.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.76 (s, 1H, phenyl-H), 7.56 (d, J = 8.1 Hz, 1H, phenyl-H), 7.41 (t, J = 8.0 Hz, 1H, phenyl-H), 7.30 (d, J = 7.7 Hz, 1H, phenyl-H), 6.86 (s, 1H, N-H H), 5.70 (s, 1H, CH-12), 4.53 (dd, J = 10.5, 6.0 Hz, 1H, CH-3), 3.66 (dd, J = 10.9, 5.4 Hz, 8H, morpholine-H), 2.83 (dt, J = 13.7, 3.7 Hz, 1H, CH-1), 2.37 (s, 1H, CH3-27), 2.34–2.25 (m, 1H, CH-9), 1.37 (s, 3H), 1.23 (s, 3H, CH3-25), 1.17 (s, 3H, CH3-26), 1.13 (s, 3H, CH3-29), 0.96 (s, 3H, CH3-23), 0.90 (s, 3H, CH3-24), 0.85 (s, 1H, CH-5), 0.82 (s, 3H, CH3-28); 13C-NMR (101 MHz, cdcl3) δ 199.98 (C11), 174.04 (C30), 169.66 (C13), 153.43 (carbamoyl), 138.71 (phenyl), 131.40 (q, J = 32.4 Hz, CF3), 129.50 (C12), 128.49 (C12), 125.23 (phenyl), 122.52 (phenyl), 121.43 (phenyl), 119.75 (phenyl), 115.21 (phenyl), 82.17 (C3), 66.95 (morpholine C), 61.67 (C9), 55.09 (C5), 48.23 (C18), 45.30 (C14), 43.80 (C20), 43.66 (morpholine C), 43.30 (C8), 38.77 (C1), 38.23 (C4), 37.69 (C22), 36.91 (C10), 33.28 (morpholine C), 32.71 (C7), 31.78 (C17), 28.42 (C28), 28.10 (C23), 26.97 (C2), 26.70 (C15), 26.42 (C16), 23.87 (C29), 23.12 (C27), 18.68 (C26), 17.36 (C6), 16.80 (C25), (C24); HRMS (m/z): [M + Na]+ calcd. for C42H58F3N2O5: 727.42978, found: 727.43507.
3β-(((3,5-bis(trifluoromethyl)phenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4k), white solid; Yield, 88.9%; m.p. 313.4–314.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.92 (s, 2H, phenyl-H), 7.52 (s, 1H, phenyl-H), 7.16 (d, J = 2.5 Hz, 1H, N-H), 5.69 (d, J = 2.5 Hz, 1H, CH-12), 4.52 (dd, J = 11.3, 5.7 Hz, 1H, CH-3), 3.68–3.64 (m, 8H, morpholine-H), 2.82 (dd, J = 13.7, 3.3 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.28 (d, J = 13.6 Hz, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.83 (s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d δ 199.96 (C11), 174.04 (C30), 169.76 (C13), 153.19 (carbamoyl), 139.73 (phenyl), 132.80 (CF3), 132.47 (CF3), 132.14 (CF3), 131.81 (CF3), 128.46 (C12), 127.16 (phenyl), 124.45 (phenyl), 121.74 (phenyl), 118.08 (phenyl), 116.35 (phenyl), 82.78 (C3), 66.93 (morpholine C), 61.62 (C9), 55.07 (C5), 48.20 (C18), 45.28 (C14), 43.79 (C20), 43.67 (morpholine C), 43.29 (C8), 38.72 (C1), 38.19 (C4), 37.67 (C22), 36.88 (C10), 33.23 (morpholine C), 32.68 (C7), 31.76 (C17), 28.41(C28), 28.09 (C23), 26.94 (C2), 26.68 (C15), 26.40 (C16), 23.81 (C29), 23.10 (C27), 18.66 (C26), 17.34 (C6), 16.77 (C25), 16.39 (C24); HRMS (m/z): [M + Na]+ calcd. for C43H56F6N2NaO5: 817.39911, found: 817.40464.
3β-(((3-methoxyphenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4l), white solid; Yield, 92.2%; m.p. 305.0–306.8 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.14 (s, 1H, phenyl-H), 6.85 (d, J = 8.1 Hz, 1H, phenyl-H), 6.62 (s, 1H, N-H), 6.60–6.57 (m, 1H, phenyl-H), 5.67 (s, 1H, CH-12), 4.49 (dd, J = 10.4, 6.1 Hz, 1H, CH-3), 3.81–3.75 (m, 3H, CH3), 3.68–3.55 (m, 8H, morpholine-H), 2.85–2.75 (m, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.31–2.22 (m, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.94 (s, 3H, CH3-23), 0.88 (s, 3H, CH3-24), 0.83 (s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.99 (C11), 174.02 (C30), 169.56 (C13), 160.22 (phenyl), 153.51 (carbamoyl), 139.33 (phenyl), 129.67 (phenyl), 128.49 (C12), 110.66 (phenyl), 109.12 (phenyl), 103.96 (phenyl), 81.68 (C3), 66.94 (morpholine C), 61.68 (C9), 55.24(CH3), 55.10 (C5), 48.22 (C18), 45.28 (C14), 43.78 (C20), 43.61(morpholine C), 43.27 (C8), 38.79 (C1), 38.23 (C4), 37.67 (C22), 36.90 (C10), 33.30 (morpholine C), 32.72 (C7), 31.77 (C17), 28.41(C28), 28.07 (C23), 26.97 (C2), 26.69 (C15), 26.40 (C16), 23.89 (C29), 23.09 (C27), 18.66 (C26), 17.35 (C6), 16.81(C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C42H60N2NaO6: 711.43491, found: 711.44018.
3β-(((4-methoxyphenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4m), white solid; Yield, 92.6%; m.p. 305.0–306.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.28 (s, 2H, phenyl-H), 6.86–6.79 (m, 2H, phenyl-H), 6.50 (s, 1H, N-H), 5.67 (d, J = 1.7 Hz, 1H, CH-12), 4.47 (t, J = 8.4 Hz, 1H, CH-3), 3.76 (d, J = 1.4 Hz, 3H, CH3), 3.70–3.55 (m, 8H, morpholine-H), 2.79 (d, J = 13.5 Hz, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.30–2.21 (m, 1H, CH-16), 1.35 (s, 3H, CH3-27), 1.21 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.11 (s, 3H, CH3-29), 0.93 (s, 3H, CH3-23), 0.86 (s, 3H, CH3-24), 0.82 (s, 1H, CH-5), 0.80 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 200.09 (C11), 174.08 (C30), 169.64 (C13), 155.74 (phenyl), 153.96 (carbamoyl), 131.14 (phenyl), 128.46 (C12), 120.34 (phenyl), 114.16 (phenyl), 81.39 (C3), 66.93 (morpholine C), 61.68 (C9), 55.48 (CH3), 55.06 (C5), 48.24 (C18), 45.30 (C14), 43.79 (C20), 43.60 (morpholine C), 43.28 (C8), 38.78 (C1), 38.25 (C4), 37.66 (C22), 36.90 (C10), 33.31 (morpholine C), 32.72 (C7), 31.77 (C17), 28.40 (C28), 28.06 (C23), 26.96 (C2), 26.69 (C15), 26.40 (C16), 23.90 (C29), 23.09 (C27), 18.66 (C26), 17.35 (C6), 16.80 (C25), 16.41 (C24); HRMS (m/z): [M + Na]+ calcd. for C42H60N2NaO6: 711.43491, found: 711.43961.
3β-(((4-(trifluoromethoxy)phenyl)carbamoyl)oxy)-30-morpholino-olean-12-ene-11,30-dione (4n), white solid; Yield, 94.0%; m.p. 308.0–310.1 °C; 1H-NMR (400 MHz, Chloroform-d) δ 7.43 (d, J = 8.5 Hz, 2H, phenyl-H), 7.16 (d, J = 8.4 Hz, 2H, phenyl-H), 6.75 (s, 1H, N-H), 5.70 (d, J = 1.9 Hz, 1H, CH-12), 4.51 (dd, J = 11.2, 5.5 Hz, 1H, CH-3), 3.67 (d, J = 5.3 Hz, 8H, morpholine-H), 2.87–2.78 (m, 1, CH-1H), 2.37 (s, 1H, CH-9), 2.29 (d, J = 12.8 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.17 (s, 3H, CH3-26), 1.13 (s, 3H, CH3-29), 0.96 (s, 3H, CH3-23), 0.89 (s, 3H, CH3-24), 0.85 (s, 1H, CH-5), 0.82 (s, 3H, CH3-28); 13C-NMR (101 MHz, Chloroform-d) δ 199.99 (C11), 174.03 (C30), 169.64 (C13), 153.54 (carbamoyl), 144.52 (phenyl), 136.85 (phenyl), 128.50 (C12), 121.85 (phenyl), 119.50 (CF3), 119.20 (phenyl), 81.97 (C3), 66.95 (morpholine C), 61.68 (C9), 55.08 (C5), 48.22 (C18), 45.30 (C14), 43.80 (C20), 43.67 (morpholine C), 43.30 (C8), 38.78 (C1), 38.25 (C4), 37.69 (C22), 36.91 (C10), 33.27 (morpholine C), 32.72 (C7), 31.78 (C17), 28.42(C28), 28.08 (C23), 26.97 (C2), 26.70 (C15), 26.42 (C16), 23.89 (C29), 23.12 (C27), 18.68 (C26), 17.36 (C6), 16.81 (C25), 16.43 (C24); HRMS (m/z): [M + Na]+ calcd. for C42H57F3N2NaO6: 765.40664, found: 765.41272.
3.2.4. 3β-(2-chloroacetyloxy)-11-oxo-olean-12-en-30-oic acid 5
A mixture of 18β–GA (0.47 g, 1.0 mmol) and chloroacetic anhydride (3.42 g, 20 mmol) was heated at 130 °C for 1 h. After the reaction was completed (monitored by by thin-layer chromatography), H2O (20 mL) was added to the cool solution, and the mixture was stirred for 30 min at the room temperature. The product was filtered off and washed with cold H2O.
A white solid; Yield, 98.0%; 280.7–281.7 °C. (literature [
44]: 260.8–261.8 °C);
1H-NMR (400 MHz, Chloroform-
d) δ 5.72 (s, 1H, CH-12), 4.61 (dd,
J = 11.8, 4.8 Hz, 1H, CH-3), 4.06 (d,
J = 2.3 Hz, 2H, CH
2-Cl), 2.83 (m, 1H, CH-1), 2.37 (s, 1H, CH-9), 2.19 (dd,
J = 13.6, 4.1 Hz, 1H, CH-16), 1.38 (m, 3H, CH
3-27), 1.23 (s, 3H, CH
3-25), 1.17 (s, 3H, CH
3-26), 1.13 (s, 3H, CH
3-29), 0.90 (s, 6H, CH
3-23/24), 0.84 (s, 3H, CH
3-28), 0.80 (m, 1H, CH-5);
13C-NMR (101 MHz, Chloroform-
d) δ 200.26 (C11), 181.51(C30), 169.59 (C13), 167.12 (acetyloxy C=O), 128.37 (C12), 83.00 (C3), 61.60 (C9), 54.94 (C5), 48.21 (C18), 45.43 (C14), 43.78 (C20), 43.18 (C8), 41.24 (C19), 40.80 (C-Cl), 38.64 (C1), 38.23 (C4), 37.67 (C22), 36.87 (C10), 32.62 (C7), 31.84 (C17), 30.87 (C21), 28.52 (C29), 28.43 (C28), 28.00 (C23), 26.43 (C2), 26.34 (C15), 23.41 (C16), 23.35 (C27), 18.64 (C26), 17.30 (C6), 16.61 (C25), 16.39 (C24); HRMS (
m/
z): [M + H]
+ calcd. for C
32H
48ClO
5: 547.3190, found: 547.3188.
3.2.5. General Procedure for Preparation of Carbamate Derivatives (6a–6d)
Compound 5 (0.55 g, 1.0 mmol), substituted secondary amine (1.5 mmol), K2CO3 (0.69 g, 5.0 mmol), and a catalytic amount of I2 in absolute ethanol (15 mL) was stirred under reflux for 12 h. The reaction mixture was evaporated under reduced pressure and the residue was dissolved in ethanol/H2O mixture and the white precipitate was collected by filtration.
3β-(2-morpholinoacetoxy)-11-oxo-olean-12-en-30-oic acid (6a), white solid; Yield, 92.0%; m.p.275.3–276.4 °C; 1H-NMR (400 MHz, Chloroform-d) δ 5.72 (t, J = 3.0 Hz, 1H, CH-12), 4.60 (dd, J = 11.2, 5.5 Hz, 1H, CH-3), 3.76 (d, J = 4.9 Hz, 4H, morpholine), 3.26–3.20 (m, 2H, CH2), 2.80 (d, J = 12.9 Hz, 1H, CH-1), 2.63 (s, 4H, morpholine), 2.37 (t, J = 3.0 Hz, 1H, CH-9), 2.19 (d, J = 13.5 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.27 (s, 3H, CH3-25), 1.16 (s, 3H, CH3-26), 1.13 (s, 3H, CH3-29), 0.87 (s, 3H, CH3-23/24), 0.83 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.25 (C11), 181.03 (C30), 169.70 (C13), 169.51 (Acetoxy, C=O), 128.40 (C12), 81.09 (C3), 66.67 (morpholine C), 61.64 (C9), 59.53 (Acetoxy, CH2), 54.93 (C5), 53.14 (morpholine C), 48.25 (C18), 45.43 (C14), 43.75 (C20), 43.19 (C8), 40.90 (C19), 38.68 (C1), 38.07 (C4), 37.70 (C22), 36.89 (C10), 32.65 (C7), 31.86 (C17), 30.93 (C21), 28.55 (C29), 28.44 (C28), 28.13 (C23), 26.46 (C2), 26.37 (C15), 23.66 (C16), 23.36 (C27), 18.66 (C26), 17.36 (C6), 16.78 (C25), 16.42 (C24); HRMS (m/z): [M + Na]+ calcd. for C36H56NO6: 598.41076, found: 598.41500.
3β-(2-(4-carbamoylpiperidin-1-yl)acetoxy)-11-oxo-olean-12-en-30-oic acid (6b), white solid; Yield, 91.1%; m.p. 283.5–285.0 °C; 1H-NMR (400 MHz, Chloroform-d) δ 5.67 (s, 1H, CH-12), 4.74 (s, 2H, NH2), 4.58 (dd, J = 11.6, 4.7 Hz, 1H, CH-3), 3.93 (t, J = 5.0 Hz, 2H, -CH2-C=O), 3.68 (dt, J = 8.8, 5.0 Hz, 4H, -CH2-N-CH2-), 3.29 (s, 2H, -CH2-C=O), 2.79 (d, J = 13.5 Hz, 1H, -CH-CONH2), 2.62 (dt, J = 11.0, 5.1 Hz, 4H, -CH2-N-CH2-), 2.35 (s, 1H, CH-9), 2.19 (dd, J = 13.4, 4.1 Hz, CH-16), 1.37 (s, 3H, CH3-27), 1.18 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.86 (s, 3H, CH3-23/24), 0.81 (s, 3H, CH3-28), 0.79 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.09 (C11), 181.10 (C30), 169.95 (C13), 169.54 (-CONH2), 169.08 (-COOC-), 128.26 (C12), 81.41 (C3), 61.60 (C9), 54.88 (C5), 53.29 (-CH2-COO-), 53.29 (-CH2-N-CH2-), 52.95 (-CH2-N-CH2-), 52.72 (-CH2-N-CH2-), 52.44 (-CH2-N-CH2-), 52.10 (-CH2-N-CH2-), 48.43 (C18), 45.39 (C14), 43.92 (C20), 43.20 (C8), 41.26 (CH-CONH2), 41.06 (C19), 38.68 (C1), 38.04 (C4), 37.88 (C22), 36.85 (C10), 32.61(C7), 31.86 (C17), 31.16 (C21), 28.66 (C29), 28.60 (C28), 28.14 (C23), 26.47 (C2), 26.39 (C16), 23.60 (C15), 23.35 (C27), 21.31[-(CH2)-CH-CONH2], 18.64 (C26), 17.34 (C6), 16.75 (C25), 16.40 (C24); HRMS (m/z): [M + Na]+ calcd. for C38H58N2NaO6: 661.41926, found: 661.41747.
3β-(2-(4-methylpiperazin-1-yl)acetoxy)-11-oxo-olean-12-en-30-oic acid (6c), white solid; Yield, 89.9%; m.p.287.5–288.9 °C; 1H-NMR (400 MHz, Chloroform-d) δ 5.70 (s, 1H, CH-12), 4.59 (dd, J = 11.7, 4.7 Hz, 1H, CH-3), 3.27 (s, 2H, -CH2-C=O), 3.14 (q, J = 7.3 Hz, 4H, 4-methylpiperazin-1-yl, CH2), 2.87 (s, 4H, 4-methylpiperazin-1-yl, CH2), 2.55 (s, 3H, CH3-piperazin-1-yl), 2.36 (s, 1H, CH-9), 2.20 (d, J = 12.0 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.13 (s, 3H, CH3-29), 0.86 (s, 3H, CH3-23/24), 0.82 (s, 3H, CH3-28), 0.79 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.08 (C11), 179.96 (C30), 169.62 (C13/ -COOC-), 128.36 (C12), 81.35 (C3), 61.61 (C9), 58.65 (-CH2-COO-), 54.91 (C5), 53.87 (-CH2-COO-), 48.37 (C18), 45.93 (4-methylpiperazin-1-yl, CH2), 45.38 (C14), 44.33 (CH3-piperazin-1-yl), 43.78 (C20), 43.18 (C8), 41.18 (C19), 38.68 (C1), 38.05 (C4), 37.80 (C22), 36.87 (C10), 32.63 (C7), 31.85 (C17), 31.10 (C21), 28.60 (C28/29), 28.51(C29), 28.13 (C23), 26.46 (C2), 26.39 (C16), 23.60 (C15), 23.35 (C27), 18.65 (C26), 17.35 (C6), 16.74 (C25), 16.40 (C24); HRMS (m/z): [M + H]+ calcd. for C37H59N2O5: 611.44240, found: 611.44228.
3β-(2-(4-(pyridin-2-yl)piperazin-1-yl)acetoxy)-11-oxo-olean-12-en-30-oic acid (6d), white solid; Yield, 88.6%; m.p. 294.0–295.6 °C; 1H-NMR (400 MHz, Chloroform-d) δ 8.21 (dd, J = 5.0, 1.8 Hz, 1H, pyridyl-H), 7.59–7.44 (m, 1H, pyridyl-H), 6.71–6.61 (m, 2H, pyridyl-H), 5.71 (s, 1H, CH-12), 4.61 (dd, J = 11.6, 4.8 Hz, 1H, CH-3), 3.63 (t, J = 5.0 Hz, 4H, piperazinyl-H), 3.30 (s, 2H, -CH2-C=O), 2.82–2.71 (m, 4H, piperazinyl-H), 2.37 (s, 1H, CH-9), 2.19 (dd, J = 13.4, 4.2 Hz, 1H, CH-16), 1.37 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.15 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.88 (s, 3H, CH3-23), 0.87 (s, 3H, CH3-24), 0.83 (s, 3H, CH3-28), 0.80 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.20 (C11), 180.64 (C30), 169.80 (C13), 169.49 (-COOC-), 158.93 (pyridin-2-yl C), 147.29 (pyridin-2-yl C), 137.93 (pyridin-2-yl C), 128.38 (C12), 113.36 (pyridin-2-yl C), 107.47(pyridin-2-yl C), 81.16 (C3), 61.62 (C9), 59.26 (-CH2-COO-), 54.92 (C5), 52.54 (piperazinyl C), 48.24 (C18), 45.41 (C14), 45.15 (piperazinyl C), 43.73 (C20), 43.17 (C8), 40.92 (C19), 38.68 (C1), 38.06 (C4), 37.68 (C22), 36.87 (C10), 32.64 (C7), 31.85 (C17), 30.93 (C21), 28.52 (C28), 28.44 (C29), 28.14 (C23), 26.44 (C2), 26.36 (C16), 23.65 (C15), 23.35 (C27), 18.64 (C26), 17.35 (C6), 16.77 (C25), 16.40 (C24); HRMS (m/z): [M + H]+ calcd. for C41H60N3O5: 674.45330, found: 674.45080.
3.2.6. General Procedure for Preparation of Carbamate Derivatives (7a–7b)
18β-GA (0.47 g, 1.0 mmol) was dissolved in ethyl acetate (15 mL) and triethylamine (0.39 g, 3.6 mmol) was added. While the mixture was stirred at room temperature, substituted acyl chloride (3.0 mmol) was added dropwise into the solution. After be stirred under reflux for 24 h, the reaction mixture was poured into water (40 mL). The organic layer was washed with 5% of aqueous NaHCO3, brine and was dried over anhydrous Na2SO4. The organic layer was then concentrated under reduced pressure. The residue obtained was purified by silica gel column chromatography using CH2Cl2–CH3OH as the eluent.
3β-(2-(4-chlorophenyl)acetoxy)-11-oxo-olean-12-en-30-oic acid (7a), white solid; Yield, 97.0%; m.p. 312.0–312.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 13.09 (s, 1H, -CH=C-OH), 7.32 (m, 2H, phenyl-H), 7.23 (m, 2H, phenyl-H), 7.04 (m, 2H, phenyl-H), 5.70 (s, 1H, CH-12), 4.76 (d, J = 7.1 Hz, 1H, -CH=C-OH), 4.46–4.40 (m, 1H, CH-3), 3.58 (s, 1H, -CH2-C=O), 3.38 (s, 1H, -CH=C-OH), 2.83–2.74 (m, 1H, CH-1), 2.35 (s, 1H, CH-9), 2.18 (d, J = 12.3 Hz, 1H, CH-16),1.36 (s, 3H, CH3-27), 1.22 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.12 (s, 3H, CH3-29), 0.83 (s, 3H, CH3-23), 0.80 (s, 3H, CH3-24), 0.75 (s, 3H, CH3-28), 0.72 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.39, 200.29, 200.23, 181.25, 173.18, 172.11, 170.87, 169.48, 134.70, 134.47, 133.20, 133.12, 132.86, 132.79, 132.65, 131.38, 130.95, 130.89, 130.85, 130.63, 130.21, 129.01, 128.96, 128.90, 128.87, 128.60, 128.55, 128.38, 104.33, 81.76, 81.28, 63.34, 61.61, 54.91, 54.84, 48.20, 48.00, 47.95, 45.41, 45.39, 43.75, 43.16, 43.14, 41.27, 40.79, 38.58, 38.23, 38.17, 38.11, 38.04, 37.66, 36.86, 36.83, 36.80, 32.60, 31.83, 30.87, 28.51, 28.41, 28.01, 27.91, 26.42, 26.34, 23.47, 23.34, 18.63, 18.61, 17.27, 16.61, 16.36, 16.33, 16.29, 16.13; HRMS (m/z): [M + Na]+ calcd. for C38H51ClNaO5: 645.33227, found: 645.33653.
3β-(2-(4- fluorophenyl)acetoxy)-11-oxo-olean-12-en-30-oic acid (7b), white solid; Yield, 85.1%; m.p. 305.0–307.7 °C; 1H-NMR (400 MHz, Chloroform-d) δ 13.05 (s, 1H, -CH=C-OH), 7.30–7.20 (m, 1H, phenyl-H), 7.15–6.77 (m, 5H, phenyl-H), 5.68 (s, 1H, CH-12), 4.76 (d, J = 8.3 Hz, 1H, -CH=C-OH), 4.58–4.42 (m, 1H, CH-3), 3.57 (s, 1H, -CH2-C=O), 3.37 (s, 1H, -CH=C-OH), 2.82–2.70 (m, 1H, CH-1), 2.32 (s, 1H, CH-9), 2.16 (d, J = 13.1 Hz, 1H, CH-16), 1.34 (s, 3H, CH3-27), 1.20 (s, 3H, CH3-25), 1.14 (s, 3H, CH3-26), 1.10 (s, 3H, CH3-29), 0.80 (s, 3H, CH3-23), 0.78 (s, 3H, CH3-24), 0.73 (s, 3H, CH3-28), 0.70 (s, 1H, CH-5); 13C-NMR (101 MHz, Chloroform-d) δ 200.35, 200.30, 200.25, 181.69, 173.49, 172.34, 171.17, 169.63, 169.56, 167.94, 167.84, 163.11, 162.96, 161.42, 161.37, 160.82, 160.73, 160.67, 160.52, 131.98, 131.38, 131.30, 131.27, 131.19, 131.14, 131.11, 131.08, 131.06, 131.03, 131.00, 130.84, 130.76, 130.70, 130.55, 130.44, 130.38, 130.36, 130.31, 130.09, 130.06, 128.76, 128.36, 128.19, 127.97, 115.88, 115.83, 115.74, 115.71, 115.67, 115.62, 115.53, 115.50, 115.46, 115.41, 115.37, 115.35, 115.32, 115.25, 115.19, 115.12, 115.11, 104.25, 82.52, 82.45, 81.63, 81.17, 63.17, 63.11, 61.61, 61.57, 54.97, 54.91, 54.83, 48.20, 47.84, 47.80, 45.42, 45.40, 43.78, 43.17, 43.14, 41.11, 40.77, 40.24, 39.94, 38.65, 38.59, 38.16, 38.11, 38.07, 38.02, 38.00, 37.66, 36.87, 36.83, 36.81, 32.60, 31.83, 30.85, 28.51, 28.43, 27.99, 27.92, 27.88, 26.42, 26.34, 23.47, 23.34, 23.32, 18.63, 18.60, 17.26, 16.59, 16.36, 16.33, 16.29, 16.24, 16.05; HRMS (m/z): [M + Na]+ calcd. for C38H51FNaO5: 629.36182, found: 629.35058.