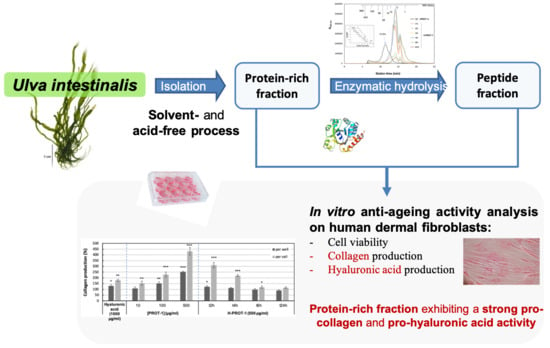
Ulva intestinalis Protein Extracts Promote In Vitro Collagen and Hyaluronic Acid Production by Human Dermal Fibroblasts
Abstract
:1. Introduction
2. Results and Discussion
2.1. Production and Characterization of Protein Fraction (PROT-1) and Protein Hydrolysates (H-PROT-1)
2.2. Assessment of PROT-1 and H-PROT-1 Cytotoxicity on NHDF
2.3. Effect of PROT-1 and H-PROT-1 on Collagen Production
2.4. Effects of PROT-1 and H-PROT-1 on Hyaluronic Acid Production
3. Material and Methods
3.1. Material
3.2. Methods
3.2.1. Protein Extraction & SEPROSYS Extraction Procedure
3.2.2. Enzymatic Hydrolysis of PROT-1 Proteins
3.2.3. Biochemical Composition
3.2.4. High-Performance Liquid Size Exclusion Chromatography
3.2.5. Ultra-High-Performance Liquid Chromatography Coupled to High Resolution Mass Spectrometry Analysis (UHPLC-HRMS)
3.2.6. SDS-PAGE
3.2.7. Cell Culture
3.2.8. Cell Viability
3.2.9. LDH Assay
3.2.10. Collagen Quantification
3.2.11. Hyaluronic Acid Quantification
3.2.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RT | room temperature |
| ECM | extracellular matric |
| HA | hyaluronic acid |
| MMP | matrix metalloproteinase |
| TIMP | tissue inhibitor of metalloproteinase |
| SM | size markers |
| HPL-SEC | high-performance liquid size-exclusion chromatography |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| LDH | lactate dehydrogenase |
| TGF-β | transforming growth factor beta |
| AP-1 | activator protein 1 |
| NF-κB | nuclear factor-kappa B |
| EDTA | ethylenediaminetetraacetic acid |
References
- Kammeyer, A.; Luiten, R.M. Oxidation events and skin aging. Ageing Res. Rev. 2015, 21, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Farage, M.A.; Miller, K.W.; Elsner, P.; Maibach, H.I. Intrinsic and extrinsic factors in skin ageing: A review. Int. J. Cosmet. Sci. 2008, 30, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Naylor, E.C.; Watson, R.E.B.; Sherratt, M.J. Molecular aspects of skin ageing. Maturitas 2011, 69, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Necas, J.; Bartosikova, L.; Brauner, P.; Kolar, J. Hyaluronic acid (hyaluronan): A review. Vet. Med. (Praha) 2008, 53, 397–411. [Google Scholar] [CrossRef] [Green Version]
- Manuskiatti, W.; Maibach, H.I. Hyaluronic acid and skin: Wound healing and aging. Int. J. Dermatol. 1996, 35, 539–544. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [Green Version]
- Boisnic, S.; Branchet, M.-C. Vieillissement cutané chronologique. EMC Dermatol. Cosmétol. 2005, 2, 232–241. [Google Scholar] [CrossRef]
- Rittié, L.; Fisher, G.J. UV-light-induced signal cascades and skin aging. Ageing Res. Rev. 2002, 1, 705–720. [Google Scholar] [CrossRef]
- Balboa, E.M.; Conde, E.; Soto, M.L.; Pérez-Armada, L.; Domínguez, H. Cosmetics from Marine Sources. In Springer Handbook of Marine Biotechnology; Kim, S.-K., Ed.; Springer Handbooks; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1015–1042. ISBN 978-3-642-53971-8. [Google Scholar]
- Couteau, C.; Coiffard, L. Chapter 14—Seaweed application in cosmetics. In Seaweed in Health and Disease Prevention; Fleurence, J., Levine, I., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 423–441. ISBN 978-0-12-802772-1. [Google Scholar]
- Fleurence, J. Seaweed proteins: Biochemical, nutritional aspects and potential uses. Trends Food Sci. Technol. 1999, 10, 25–28. [Google Scholar] [CrossRef]
- van den Burg, S.; Stuiver, M.; Veenstra, F.; Bikker, P.; López-Contreras, A.; Palstra, A.; Broeze, J.; Jansen, H.; Jak, R.; Gerritsen, A.; et al. A Triple P Review of the Feasibility of Sustainable Offshore Seaweed Production in the North Sea; Wageningen University & Research Centre: Wageningen, The Netherlands, 2013; ISBN 978-90-8615-652-8. [Google Scholar]
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Chandini, S.K.; Ganesan, P.; Suresh, P.V.; Bhaskar, N. Seaweeds as a source of nutritionally beneficial compounds—A review. J. Food Sci. Technol. 2008, 45, 1–13. [Google Scholar]
- Silva, M.; Vieira, L.; Almeida, A.P.; Kijjoa, A. The marine macroalgae of the genus Ulva: Chemistry, biological activities and potential applications. J. Oceanogr. Mar. Res. 2013, 1, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Adrien, A.; Bonnet, A.; Dufour, D.; Baudouin, S.; Maugard, T.; Bridiau, N. Pilot production of ulvans from Ulva sp. and their effects on hyaluronan and collagen production in cultured dermal fibroblasts. Carbohydr. Polym. 2017, 157, 1306–1314. [Google Scholar] [CrossRef]
- Adrien, A.; Dufour, D.; Baudouin, S.; Maugard, T.; Bridiau, N. Evaluation of the anticoagulant potential of polysaccharide-rich fractions extracted from macroalgae. Nat. Prod. Res. 2017, 31, 2126–2136. [Google Scholar] [CrossRef]
- Benjama, O.; Masniyom, P. Nutritional composition and physicochemical properties of two green seaweeds (Ulva pertusa and U. intestinalis) from the Pattani Bay in Southern Thailand. Songklanakarin J. Sci. Technol. 2011, 33, 575–583. [Google Scholar]
- Majmudar, G. Compositions of Marine Botanicals to Provide Nutrition to Aging and Environmentally Damaged Skin. U.S. Patent No. 7303753B2, 4 December 2007. [Google Scholar]
- Baudouin, S.; Dufour, D.; Yao, J. Treating Algae by Diffusing Algae in Water, Filtering Pulp, Ultrafiltration of Pressing Juice, Demineralizing Ultrafiltration Retentate and Decanting Demineralized Retentate, and Recovering Vegetable Protein and Sulfated Polysaccharide. F.R. Patent No. 2998894B1, 31 July 2015. [Google Scholar]
- Shuuluka, D.; Bolton, J.J.; Anderson, R.J. Protein content, amino acid composition and nitrogen-to-protein conversion factors of Ulva rigida and Ulva capensis from natural populations and Ulva lactuca from an aquaculture system, in South Africa. J. Appl. Phycol. 2013, 25, 677–685. [Google Scholar] [CrossRef]
- Wijesekara, I.; Lang, M.; Marty, C.; Gemin, M.-P.; Boulho, R.; Douzenel, P.; Wickramasinghe, I.; Bedoux, G.; Bourgougnon, N. Different extraction procedures and analysis of protein from Ulva sp. in Brittany, France. J. Appl. Phycol. 2017, 29, 2503–2511. [Google Scholar] [CrossRef]
- Lewis, E.J.; Gonzalves, E.A. Amino acid contents of some marine algae from Bombay. New Phytol. 1960, 59, 109–115. [Google Scholar] [CrossRef]
- Lewis, E.J.; Gonzalves, E.A. Periodic studies of the proteins, peptides, and free amino-acids in Enteromorpha prolifera f. capillaris and Ulva lactuca var. rigida. Ann. Bot. 1962, 26, 318–327. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Wu, H.; Liu, R. Overview on biological activities and molecular characteristics of sulfated polysaccharides from marine green algae in recent years. Mar. Drugs 2014, 12, 4984–5020. [Google Scholar] [CrossRef]
- Abdel-fattah, A.F.; Sary, H.H. Glycoproteins from Ulva lactuca. Phytochemistry 1987, 26, 1447–1448. [Google Scholar] [CrossRef]
- Rouxel, C.; Bonnabeze, E.; Daniel, A.; Jérôme, M.; Etienne, M.; Fleurence, J. Identification by SDS PAGE of green seaweeds (Ulva and Enteromorpha) used in the food industry. J. Appl. Phycol. 2001, 13, 215–218. [Google Scholar] [CrossRef]
- Girardeau-Hubert, S.; Teluob, S.; Pageon, H.; Asselineau, D. The reconstructed skin model as a new tool for investigating in vitro dermal fillers: Increased fibroblast activity by hyaluronic acid. Eur. J. Dermatol. 2015, 25, 312–322. [Google Scholar] [CrossRef]
- Mast, B.A.; Diegelmann, R.F.; Krummel, T.M.; Cohen, I.K. Hyaluronic acid modulates proliferation, collagen and protein synthesis of cultured fetal fibroblasts. Matrix 1993, 13, 441–446. [Google Scholar] [CrossRef]
- Schwach-Abdellaoui, K.; Malle, B.M. Compositions with Several Hyaluronic Acid Fractions for Cosmetic Use. W.O. Patent No. 2008003321A3, 13 March 2008. [Google Scholar]
- Turley, E. Topically Administered, Skin-Penetrating Glycosaminoglycan Formulations Suitable for Use in Cosmetic and Pharmaceutical Applications. W.O. Patent No. 2011140630A1, 17 November 2011. [Google Scholar]
- Kmail, A.; Lyoussi, B.; Zaid, H.; Saad, B. In vitro assessments of cytotoxic and cytostatic effects of Asparagus aphyllus, Crataegus aronia, and Ephedra alata in monocultures and co-cultures of Hepg2 and THP-1-derived macrophages. Pharmacogn. Commun. 2015, 5, 165–172. [Google Scholar] [CrossRef] [Green Version]
- Ko, H.J.; Kim, G.B.; Lee, D.H.; Lee, G.S.; Pyo, H.B. The effect of hydrolyzed Jeju Ulva pertusa on the proliferation and type I collagen synthesis in replicative senescent fibroblasts. J. Soc. Cosmet. Sci. Korea 2013, 39, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Montanari, D.; Guglielmo, M. Cosmetic Composition with a Lifting Effect for Sustaining Relaxed Skin tissues. W.O. Patent No. 2008146116A2, 4 December 2008. [Google Scholar]
- Honma, Y.; Kikuchi, K.; Uemura, H.; Inaoka, S.; Tsunetsugu, S. Peptides that Increase Collagen or Hyaluronic acid Production. U.S. Patent No. 7989590B2, 2 August 2011. [Google Scholar]
- Joe, M.-J.; Kim, S.-N.; Choi, H.-Y.; Shin, W.-S.; Park, G.-M.; Kang, D.-W.; Kim, Y.K. The inhibitory effects of eckol and dieckol from Ecklonia stolonifera on the expression of matrix metalloproteinase-1 in human dermal fibroblasts. Biol. Pharm. Bull. 2006, 29, 1735–1739. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Liou, S.-F.; Chen, S.-J.; Shih, M.-F. Protective effects of Chlorella-derived peptide on UVB-induced production of MMP-1 and degradation of procollagen genes in human skin fibroblasts. Regul. Toxicol. Pharmacol. 2011, 60, 112–119. [Google Scholar] [CrossRef]
- Kim, C.-R.; Kim, Y.-M.; Lee, M.-K.; Kim, I.-H.; Choi, Y.-H.; Nam, T.-J. Pyropia yezoensis peptide promotes collagen synthesis by activating the TGF-β/Smad signaling pathway in the human dermal fibroblast cell line Hs27. Int. J. Mol. Med. 2017, 39, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, R.D.; Berry, M.G.; Navsaria, H.A. Hyaluronic acid: The scientific and clinical evidence. J. Plast. Reconstr. Aesthet. Surg. 2007, 60, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Tanimoto, K.; Suzuki, A.; Ohno, S.; Honda, K.; Tanaka, N.; Doi, T.; Yoneno, K.; Ohno-Nakahara, M.; Nakatani, Y.; Ueki, M.; et al. Effects of TGF-β on hyaluronan anabolism in fibroblasts derived from the synovial membrane of the rabbit temporomandibular joint. J. Dent. Res. 2004, 83, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Fayad, S.; Nehmé, R.; Tannoury, M.; Lesellier, E.; Pichon, C.; Morin, P. Macroalga Padina pavonica water extracts obtained by pressurized liquid extraction and microwave-assisted extraction inhibit hyaluronidase activity as shown by capillary electrophoresis. J. Chromatogr. A 2017, 1497, 19–27. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef]
- Bremner, J.M. Determination of nitrogen in soil by the Kjeldahl method. J. Agric. Sci. 1960, 55, 11–33. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144. [Google Scholar]
- Chabrol, E.; Charonnat, R. Une nouvelle réaction pour l’étude des lipides: L’oléidemie. Presse Méd. 1937, 45, 1713–1714. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Legrand, C.; Bour, J.M.; Jacob, C.; Capiaumont, J.; Martial, A.; Marc, A.; Wudtke, M.; Kretzmer, G.; Demangel, C.; Duval, D.; et al. Lactate dehydrogenase (LDH) activity of the number of dead cells in the medium of cultured eukaryotic cells as marker. J. Biotechnol. 1992, 25, 231–243. [Google Scholar] [CrossRef]
- Tullberg-Reinert, H.; Jundt, G. In situ measurement of collagen synthesis by human bone cells with a Sirius Red-based colorimetric microassay: Effects of transforming growth factor β2 and ascorbic acid 2-phosphate. Histochem. Cell Biol. 1999, 112, 271–276. [Google Scholar] [CrossRef]
- Dreno, B.; Araviiskaia, E.; Berardesca, E.; Bieber, T.; Hawk, J.; Sanchez-Viera, M.; Wolkenstein, P. The science of dermocosmetics and its role in dermatology. J. Eur. Acad. Dermatol. Venereol. JEADV 2014, 28, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the PROT-1 and H-PROT-1 fractions are available from the authors. |











| Fraction | Ashes | Proteins | Neutral Sugars | Uronic Acids | Lipids | Polyphenols |
|---|---|---|---|---|---|---|
| % (w/wdry extract) | ||||||
| PROT-1 | 20 | 51 | 22 | 6 | <1 | <1 |
| Entry | Retention Time (min) | Ion | Experimental Monoisotopic Mass (Da) | Mass Accuracy (ppm) | Peptide or Amino Acid |
|---|---|---|---|---|---|
| 1 | 2.58 | [M + H]+ | 233.1130 | 0.86 | VD |
| 2 | 3.20 | [M + H]+ | 508.1899 | 2.56 | EGESS |
| 3 | 3.81 | [M + H]+ | 247.1284 | 2.02 | VE |
| 4 | 4.16 | [M + H]+ | 246.1456 | 2.84 | I/LN |
| 5 | 5.65 | [M + H]+ | 247.1289 | 2.02 | I/LD |
| 6 | 5.65 | [M + H]+ | 247.1294 | 2.02 | EV |
| 7 | 6.67 | [M + H]+ | 294.1545 | 2.72 | I/L-hexose |
| 8 | 8.21 | [M + H]+ | 261.1452 | 0.77 | I/LE |
| 9 | 8.81 | [M + H]+ | 261.1452 | 0.77 | I/LE |
| 10 | 9.57 | [M + H]+ | 590.2798 | 2.88 | ETVNQ |
| 11 | 9.80 | [M + H]+ | 404.2132 | 1.98 | I/LSAN |
| 12 | 10.40 | [M + H]+ | 419.2135 | 0.00 | TVAE |
| 13 | 10.70 | [M + H]+ | 449.2243 | 1.56 | TEVT |
| 14 | 10.93 | [M + H]+ | 328.1394 | 0.61 | F-hexose |
| 15 | 11.32 | [M + H]+ | 418.2293 | 0.96 | I/LSAQ |
| 16 | 11.52 | [M + H]+ | 555.2410 | 0.18 | TGYTN |
| 17 | 11.52 | [M + H]+ | 203.14 | 4.43 | AI/L |
| 18 | 11.83 | [M + H]+ | 546.2520 | 0.18 | DAI/LNN |
| 19 | 12.46 | [M + H]+ | 502.2625 | 1.00 | AI/LNGQ |
| 20 | 12.66 | [M + H]+ | 295.1294 | 1.69 | FE |
| 21 | 12.87 | [M + H]+ | 475.2515 | 0.84 | AI/LSNA |
| 22 | 13.41 | [M + H]+ | 367.1506 | 0.27 | W-hexose |
| 23 | 14.27 | [M + H]+ | 350.1740 | 1.43 | I/LMS |
| 24 | 14.48 | [M + H]+ | 526.2163 | 4.18 | SAYDA |
| 25 | 14.48 | [M + H]+ | 526.2163 | 4.18 | * DADA |
| 26 | 15.29 | [M + H]+ | 440.1667 | 0.68 | EEY |
| 27 | 16.18 | [M + H]+ | 403.2188 | 0.07 | DVVA |
| 28 | 16.65 | [M + H]+ | 237.1241 | 3.08 | AF |
| 29 | 17.22 | [M + H]+ | 205.0982 | 2.44 | W |
| 30 | 17.60 | [M + H]+ | 229.1552 | 2.18 | I/LP |
| 31 | 18.01 | [M + H]+ | 492.2662 | 0.61 | SI/LTTA |
| 32 | 19.09 | [M + H]+ | 458.2607 | 0.59 | I/LPQT |
| 33 | 19.09 | [M + H]+ | 458.2607 | 0.59 | QPI/LT |
| 34 | 19.98 | [M + H]+ | 295.1659 | 2.03 | I/LY |
| 35 | 21.39 | [M + H]+ | 302.2077 | 0.66 | I/LI/LG |
| 36 | 21.76 | [M + H]+ | 416.2517 | 1.20 | * GSI/LA |
| 37 | 21.76 | [M + H]+ | 416.2517 | 1.20 | NVI/LA |
| 38 | 22.01 | [M + H]+ | 409.2078 | 1.00 | VGAY |
| 39 | 22.01 | [M + H]+ | 409.2078 | 1.00 | I/LNY |
| 40 | 22.25 | [M + H]+ | 552.2316 | 1.81 | EAEFG |
| 41 | 22.64 | [M + H]+ | 411.7227 | 6.07 | YTI/LDPP * |
| 42 | 22.64 | [M + H]+ | 295.1654 | 0.34 | YI/L |
| 43 | 23.03 | [M + 2H]2+ | 720.3186 | 0.99 | QAAEAAESGDFKSE |
| 44 | 23.25 | [M + 2H]2+ | 841.8622 | - | * QSDWSEAEAAHS |
| 45 | 23.45 | [M + H]+ | 777.3397 | 0.40 | NQAAEAAESGDESKF |
| 46 | 23.65 | [M + H]+ | 459.2179 | 3.64 | EEYTFTK |
| 47 | 23.65 | [M + H]+ | 459.2179 | 3.64 | KFTTYEE |
| 48 | 23.65 | [M + H]+ | 459.2179 | 3.64 | KFYTTEE |
| 49 | 23.85 | [M + H]+ | 550.2878 | 1.13 | (I/L)GTYP |
| 50 | 24.12 | [M + H]+ | 790.4323 | 2.21 | TI/LATI/LSNA |
| 51 | 24.32 | [M + H]+ | 329.1500 | 1.28 | YF |
| 52 | 24.53 | [M + H]+ | 373.2276 | 3.22 | KDTVI/LI/LG |
| 53 | 24.53 | [M + 2H]2+ | 766.8573 | - | * KDI/LI/LI/L * |
| 54 | 25.18 | [M + H]+ | 401.2756 | 0.72 | VVAI/L |
| 55 | 25.18 | [M + H]+ | 600.3360 | 1.33 | SPGGI/LGI/L |
| 56 | 25.18 | [M + H]+ | 600.3360 | 1.33 | SNI/LPGI/L |
| 57 | 25.58 | [M + H]+ | 873.3639 | 1.57 | (YG)YGDI/LDA |
| 58 | 25.58 | [M + H]+ | 873.3639 | 1.57 | (YG)GI/LDYDA |
| 59 | 25.58 | [M + H]+ | 873.3639 | 1.57 | (YG)DI/LGYDA |
| 60 | 25.58 | [M + H]+ | 873.3639 | 1.57 | (YG)DYGI/LDA |
| 61 | 25.58 | [M + H]+ | 873.3639 | 1.57 | (DI/L)GYGYDA |
| 62 | 25.86 | [M + H]+ | 687.2986 | 0.17 | DI/LGYGY |
| 63 | 26.07 | [M + H]+ | 579.3143 | 0.98 | NVI/LAY |
| 64 | 26.07 | [M + 2H]2+ | 590.7799 | 3.71 | QEYENI/LI/LGSQ |
| 65 | 26.07 | [M + H]+ | 555.2821 | 1.31 | YI/LYP |
| 66 | 26.73 | [M + H]+ | 431.2497 | 0.86 | DI/LAI/L |
| 67 | 27.04 | [M + H]+ | 401.2756 | 0.72 | I/LVGI/L |
| 68 | 24.69 | [M + 2H]2+ | 604.7886 | 7.87 | DSTWI/LTTAI/LSN |
| 69 | 28.26 | [M + 2H]2+ | 566.2830 | 5.14 | SG(Q/AG)QEI/LI/LDI/LE |
| 70 | 28.26 | [M + 2H]2+ | 566.2830 | 5.14 | QSGQEI/LI/LDI/LE |
| 71 | 28.26 | [M + 2H]2+ | 566.2830 | 5.14 | TNGQEI/LI/LDI/LE |
| 72 | 28.46 | [M + H]+ | 726.4041 | 1.13 | EI/LSPPI/LA |
| 73 | 28.46 | [M + H]+ | 726.4041 | 1.13 | ESI/LPPI/LA |
| 74 | 28.46 | [M + H]+ | 726.4041 | 1.13 | DTI/LPAI/LP |
| 75 | 30.26 | [M + H]+ | 577.3329 | 2.72 | AFI/LI/LN |
| 76 | 30.47 | [M + H]+ | 645.4180 | 4.34 | TI/LI/LVI/LS |
| 77 | 30.47 | [M + H]+ | 645.4180 | 4.34 | I/LI/LTVI/LS |
| 78 | 30.47 | [M + H]+ | 645.4180 | 4.34 | VI/LTI/LI/LS |
| 79 | 30.88 | [M + H]+ | 938.4491 | 2.66 | EI/LTDTVNF |
| 80 | 30.88 | [M + H]+ | 938.4491 | 2.66 | ETI/LDTVNF |
| 81 | 31.35 | [M + 2H]2+ | 640.3069 | 4.08 | EGESSI/LTTAI/LSNA |
| 82 | 31.35 | [M + 2H]2+ | 640.3069 | 4.08 | E(SG/DG/TG)ESI/LTTAI/LSNA |
| 83 | 31.89 | [M + 2H]2+ | 616.3179 | 6.44 | (AP)EI/LI/LTVDYNP |
| 84 | 31.89 | [M + 2H]2+ | 616.3179 | 6.44 | PVVDI/LTVDYNP |
| 85 | 32.13 | [M + H]+ | 558.3867 | 0.95 | I/LTVI/LI/L |
| 86 | 32.50 | [M + H]+ | 746.4453 | 0.74 | PI/LI/LGI/LYA |
| 87 | 32.70 | [M + H]+ | 890.4997 | - | * I/LTI/LP |
| 88 | 32.70 | [M + H]+ | 890.4997 | - | * YEI/LP |
| 89 | 32.70 | [M + H]+ | 675.4078 | 0.25 | I/LPGI/LI/LY |
| 90 | 32.70 | [M + H]+ | 675.4078 | 0.25 | I/LPI/LGI/LY |
| 91 | 33.17 | [M + 2H]2+ | 634.3411 | 4.10 | SI/LDI/LPHI/LPTQF |
| 92 | 33.37 | [M + 2H]2+ | 691.8536 | 3.45 | SI/LDI/LDPHI/LPTQF |
| Total amino Acid Composition Determination | Elucidation of Peptide Sequences | |
|---|---|---|
| UHPLC Parameters | ||
| Column | Acquity UPLC HSST3 (150 mm × 2.1 mm × 1.7 μm) maintained at 25 °C | |
| Flow rate (μL.min−1) | 0.3 mL·min−1 | |
| Gradient used for the total amino acid composition determination: water/methanol 95:5 (v/v) (A)/water/methanol 50:50 (v/v) (B) + 0.5% (v/v) formic acid Gradient used for the elucidation of peptide sequences: water (A)/acetonitrile (B) + 0.1% (v/v) formic acid | 0 min: 100% A; 2 min: 100% A; 7 min: 30% A; 8 min: 100% A; 13 min: 100% A. | 0 min: 100% A; 2 min: 100% A; 2.5 min: 99% A; 3 min: 99% A; 3.5 min: 98% A; 4 min: 97% A; 5.5 min: 97% A; 8.5 min: 95% A; 10 min: 95% A; 15 min: 90% A; 18 min: 90% A; 22 min: 80% A; 27 min: 80% A; 30 min: 70% A; 34 min: 70% A; 36 min: 0% A; 38 min: 0% A; 40 min: 100% A; 45 min: 100% A. |
| Injection | 5 μL (4 °C) | |
| MS Parameters | ||
| Mode | ESI+ MSE, centroid: - Function 1, low energy, 5 eV - Function 2, high energy, ramping from 10 to 30 eV | ESI+ MS/MS DDA, centroid: - Transition from MS to MS/MS and MS/MS to MS when ion intensity becomes higher than 500,000/s and lower than 10,000/s (or after 10 s), respectively - Collision energy ramping from 15 to 35 eV |
| Source temperature | 120 °C | |
| Desolvation temperature | 500 °C | |
| Gas flow rate of the cone | 50 L/h | |
| Desolvation gas flow rate | 300 L/h | |
| Capillary voltage | 3 kV | |
| Sampling cone voltage | 35 V | |
| Source offset | 80 V | |
| Acquisition mass range | 50–1200 m/z (0.5 scans/s) | 50–2500 m/z (0.5 scans/s) |
| Lock-mass | Leucine Enkephaline (MW = 555.62 Da,1 ng/μL) with a flow-rate of 5 μL/min | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodin, J.; Adrien, A.; Bodet, P.-E.; Dufour, D.; Baudouin, S.; Maugard, T.; Bridiau, N. Ulva intestinalis Protein Extracts Promote In Vitro Collagen and Hyaluronic Acid Production by Human Dermal Fibroblasts. Molecules 2020, 25, 2091. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25092091
Bodin J, Adrien A, Bodet P-E, Dufour D, Baudouin S, Maugard T, Bridiau N. Ulva intestinalis Protein Extracts Promote In Vitro Collagen and Hyaluronic Acid Production by Human Dermal Fibroblasts. Molecules. 2020; 25(9):2091. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25092091
Chicago/Turabian StyleBodin, Justine, Amandine Adrien, Pierre-Edouard Bodet, Delphine Dufour, Stanislas Baudouin, Thierry Maugard, and Nicolas Bridiau. 2020. "Ulva intestinalis Protein Extracts Promote In Vitro Collagen and Hyaluronic Acid Production by Human Dermal Fibroblasts" Molecules 25, no. 9: 2091. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25092091