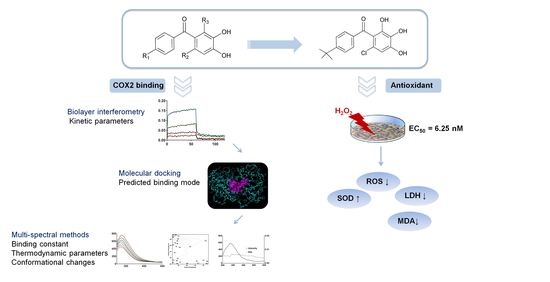
An Investigation into the Interaction between Double Hydroxide-Based Antioxidant Benzophenone Derivatives and Cyclooxygenase 2
Abstract
:1. Introduction
2. Results
2.1. Biolayer Interferometry Studies
2.2. Molecular Docking Studies
2.3. Fluorescence Spectral Studies
2.3.1. Fluorescence Quenching Mechanisms
2.3.2. Binding Constants and Number of Binding Sites
2.3.3. Thermodynamic Parameters
2.3.4. Fluorescence Resonance Energy Transfer Studies
2.3.5. Conformational Studies
2.4. Biological Activities
2.4.1. In Vitro Antioxidant Activity
2.4.2. Cytotoxocity of Compound SC2
2.4.3. Effects of Oxidative Stress-Related Factors
3. Discussion
4. Materials and Methods
4.1. Biolayer Interferometry Studies
4.2. Molecular Docking
4.3. Fluorescence Spectrum Measurement
4.4. Pharmacological/Biological Assays
4.4.1. Cell Lines and Cell Culture
4.4.2. Establishment of the Oxidative Damage Model
4.4.3. Protective Effect of Target Compounds on Oxidative Damage
4.4.4. Activity Assessment of LDH, MDA, SOD and ROS In Vitro
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Mitchell, J.A.; Kirkby, N.S.; Ahmetaj-Shala, B.; Armstrong, P.C.; Crescente, M.; Ferreira, P.; Lopes Pires, M.E.; Vaja, R.; Warner, T.D. Cyclooxygenases and the cardiovascular system. Pharmacol. Ther. 2021, 217, 107624. [Google Scholar] [CrossRef]
- Sağlık, B.N.; Osmaniye, D.; Levent, S.; Çevik, U.A.; Çavuşoğlu, B.K.; Özkay, Y.; Kaplancıklı, Z.A. Design, synthesis and biological assessment of new selective COX-2 inhibitors including methyl sulfonyl moiety. Eur. J. Med. Chem. 2021, 209, 112918. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Diwan, A.; Thabet, H.K.H.; Imran, M.; Bakht, A. Discovery of Novel Pyridazine-Based Cyclooxygenase-2 Inhibitors with a Promising Gastric Safety Profile. Molecules 2020, 25, 2002. [Google Scholar] [CrossRef] [PubMed]
- Hashemi Goradel, N.; Najafi, M.; Salehi, E.; Farhood, B.; Mortezaee, K. Cyclooxygenase-2 in cancer: A review. J. Cell. Physiol. 2019, 234, 5683–5699. [Google Scholar] [CrossRef] [PubMed]
- Szczukowski, Ł.; Krzyżak, E.; Zborowska, A.; Zając, P.; Potyrak, K.; Peregrym, K.; Wiatrak, B.; Marciniak, A.; Świątek, P. Design, Synthesis and Comprehensive Investigations of Pyrrolo[3,4-d]pyridazinone-Based 1,3,4-Oxadiazole as New Class of Selective COX-2 Inhibitors. Int. J. Mol. Sci. 2020, 21, 9623. [Google Scholar] [CrossRef] [PubMed]
- El-Shoukrofy, M.S. Pyrazoles containing thiophene, thienopyrimidine and thienotriazolopyrimidine as COX-2 selective inhibitors: Design, synthesis, in vivo anti-inflammatory activity, docking and in silico chemo-informatic studies. Bioorg. Chem. 2019, 85, 45. [Google Scholar] [CrossRef] [PubMed]
- Taher, E.S.; Ibrahim, T.S.; Fares, M.; AL-Mahmoudy, A.M.M.; Radwan, A.F.; Orabi, K.Y.; El-Sabbagh, O.I. Novel benzenesulfonamide and 1,2-benzisothiazol-3(2H)-one-1,1-dioxide derivatives as potential selective COX-2 inhibitors. Eur. J. Med. Chem. 2019, 171, 372–382. [Google Scholar] [CrossRef]
- Grosser, T.; Ricciotti, E.; FitzGerald, G.A. The Cardiovascular Pharmacology of Nonsteroidal Anti-inflammatory Drugs. Trends Pharmacol. Sci. 2017, 38, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, B.M.; Fanelli, A.; Piccinino, D.; De Angelis, M.; Dolfa, C.; Palamara, A.T.; Nencioni, L.; Zippilli, C.; Crucianelli, M.; Saladino, R. Synthesis of Stilbene and Chalcone Inhibitors of Influenza A Virus by SBA-15 Supported Hoveyda-Grubbs Metathesis. Catalysts 2019, 9, 983. [Google Scholar] [CrossRef] [Green Version]
- Karataş, M.O.; Günal, S.; Mansur, A.; Alıcı, B.; Özdemir, İ. Catechol-bearing imidazolium and benzimidazolium chlorides as promising antimicrobial agents. Arch. Pharm. 2020, 353, e2000013. [Google Scholar] [CrossRef] [PubMed]
- Razaviamri, S.; Wang, K.; Liu, B.; Lee, B.P. Catechol-Based Antimicrobial Polymers. Molecules 2021, 26, 559. [Google Scholar] [CrossRef] [PubMed]
- Jakovljević, K.; Joksović, M.D.; Matić, I.Z.; Petrović, N.; Stanojković, T.; Sladić, D.; Vujčić, M.; Janović, B.; Joksović, L.; Trifunović, S.; et al. Novel 1,3,4-thiadiazole-chalcone hybrids containing catechol moiety: Synthesis, antioxidant activity, cytotoxicity and DNA interaction studies. MedChemComm 2018, 9, 1679–1697. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Ramírez, A.; Mascayano-Collado, C.; Barriga, A.; Echeverría, J.; Urzúa, A. Inhibition of Soybean 15-Lipoxygenase and Human 5-Lipoxygenase by Extracts of Leaves, Stem Bark, Phenols and Catechols Isolated From Lithraea caustica (Anacardiaceae). Front. Pharmacol. 2020, 11, 594257. [Google Scholar] [CrossRef]
- Espíndola, K.M.M.; Ferreira, R.G.; Narvaez, L.E.M.; Silva Rosario, A.C.R.; da Silva, A.H.M.; Silva, A.G.B.; Vieira, A.P.O.; Monteiro, M.C. Chemical and Pharmacological Aspects of Caffeic Acid and Its Activity in Hepatocarcinoma. Front. Oncol. 2019, 9, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinhatti, A.V.; de Barros, F.M.; de Farias, C.B.; Schwartsmann, G.; von Poser, G.L.; Abujamra, A.L. Antiproliferative activity of the dimeric phloroglucinol and benzophenone derivatives of Hypericum spp. native to southern Brazil. Anticancer Drugs 2013, 24, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Q.; Xu, Q.; Luo, J.; Wang, L.-J.; Jiang, B.; Zhang, R.-S.; Shi, D.-Y. Design, synthesis and biological evaluation of uncharged catechol derivatives as selective inhibitors of PTP1B. Eur. J. Med. Chem. 2017, 136, 348–359. [Google Scholar] [CrossRef]
- Muhammed, K.; Manohar, S.; Husain, M. Mechanisms underlying apathy in Parkinson’s disease. Lancet Lond. Engl. 2015, 385 (Suppl 1), S71. [Google Scholar] [CrossRef]
- Haddad, F.; Sawalha, M.; Khawaja, Y.; Najjar, A.; Karaman, R. Dopamine and Levodopa Prodrugs for the Treatment of Parkinson’s Disease. Molecules 2018, 23, 40. [Google Scholar] [CrossRef] [Green Version]
- Glass, C.K.; Saijo, K.; Winner, B. Mechanisms underlying inflammation in neurodegeneration. Cell 2010, 140, 918–934. [Google Scholar] [CrossRef] [Green Version]
- Mehrabadi, S.; Sadr, S.S. Administration of Vitamin D3 and E supplements reduces neuronal loss and oxidative stress in a model of rats with Alzheimer’s disease. Neurol. Res. 2020, 42, 862–868. [Google Scholar] [CrossRef]
- Zhao, H.; Wei, D.; Li, M.; Du, Y. Substituent contribution to the genotoxicity of benzophenone-type UV filters. Ecotoxicol. Environ. Saf. 2013, 95, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, L.; Chen, X.; Niu, S.; Ban, S.; Feng, X.; Li, Q. Synthesis, In Vitro Anti-Inflammatory Activity and Molecular Docking of Butyrate Benzophenone Compound. ChemistrySelect 2019, 4, 171–174. [Google Scholar] [CrossRef]
- Yang, L.; Shi, H.; Li, Y.L.; Chen, X.; Niu, S.Q.; Qiao, X.-Z.; Mai, J.-Q.; Li, Q.-S. Synthesis and anti-inflammatory activity of novel isobutyl benzophenone derivatives. Yaoxue Xuebao 2018, 53, 256–262. [Google Scholar] [CrossRef]
- Zhao, W.; Feng, X.; Ban, S.; Lin, W.; Li, Q. Synthesis and biological activity of halophenols as potent antioxidant and cytoprotective agents. Bioorg. Med. Chem. Lett. 2010, 20, 4132–4134. [Google Scholar] [CrossRef]
- Ottosen, E.R.; Sørensen, M.D.; Björkling, F.; Skak-Nielsen, T.; Fjording, M.S.; Aaes, H.; Binderup, L. Synthesis and structure-activity relationship of aminobenzophenones. A novel class of p38 MAP kinase inhibitors with high antiinflammatory activity. J. Med. Chem. 2003, 46. [Google Scholar] [CrossRef]
- Zabiulla; Gulnaz, A.R.; Mohammed, Y.H.E.; Khanum, S.A. Design, synthesis and molecular docking of benzophenone conjugated with oxadiazole sulphur bridge pyrazole pharmacophores as anti inflammatory and analgesic agents. Bioorg. Chem. 2019, 92, 103220. [Google Scholar] [CrossRef]
- Kober, D.L.; Stuchell-Brereton, M.D.; Kluender, C.E.; Dean, H.B.; Strickland, M.R.; Steinberg, D.F.; Nelson, S.S.; Baban, B.; Holtzman, D.M.; Frieden, C.; et al. Functional insights from biophysical study of TREM2 interactions with apoE and Aβ1-42. Alzheimers Dement. J. Alzheimers Assoc. 2020. [Google Scholar] [CrossRef]
- Kaur, H.; Shorie, M.; Sabherwal, P. Biolayer interferometry-SELEX for Shiga toxin antigenic-peptide aptamers & detection via chitosan-WSe2 aptasensor. Biosens. Bioelectron. 2020, 167, 112498. [Google Scholar] [CrossRef]
- Cui, X.; Song, M.; Liu, Y.; Yuan, Y.; Huang, Q.; Cao, Y.; Lu, F. Identifying conformational changes of aptamer binding to theophylline: A combined biolayer interferometry, surface-enhanced Raman spectroscopy, and molecular dynamics study. Talanta 2020, 217, 121073. [Google Scholar] [CrossRef]
- Bandgar, B.P.; Kinkar, S.N.; Chavan, H.V.; Jalde, S.S.; Shaikh, R.U.; Gacche, R.N. Synthesis and biological evaluation of asymmetric indole curcumin analogs as potential anti-inflammatory and antioxidant agents. J. Enzyme Inhib. Med. Chem. 2014, 29, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Bhat, M.A.; Al-Omar, M.A.; Alsaif, N.A.; Almehizia, A.A.; Naglah, A.M.; Razak, S.; Khan, A.A.; Ashraf, N.M. Novel sulindac derivatives: Synthesis, characterisation, evaluation of antioxidant, analgesic, anti-inflammatory, ulcerogenic and COX-2 inhibition activity. J. Enzyme Inhib. Med. Chem. 2020, 35, 921–934. [Google Scholar] [CrossRef]
- Law, B.Y.K.; Gordillo-Martínez, F.; Qu, Y.Q.; Zhang, N.; Xu, S.W.; Coghi, P.S.; Fai Mok, S.W.; Guo, J.; Zhang, W.; Leung, E.L.H.; et al. Thalidezine, a novel AMPK activator, eliminates apoptosis-resistant cancer cells through energy-mediated autophagic cell death. Oncotarget 2017, 8, 30077–30091. [Google Scholar] [CrossRef]
- Smith, W.L.; DeWitt, D.L.; Garavito, R.M. Cyclooxygenases: Structural, Cellular, and Molecular Biology. Annu. Rev. Biochem. 2000, 69, 145–182. [Google Scholar] [CrossRef] [Green Version]
- Olomola, T.O.; Mphahlele, M.J.; Gildenhuys, S. Benzofuran-selenadiazole hybrids as novel α-glucosidase and cyclooxygenase-2 inhibitors with antioxidant and cytotoxic properties. Bioorg. Chem. 2020, 100, 103945. [Google Scholar] [CrossRef] [PubMed]
- Atatreh, N. Anti-inflammatory Drug Approach: Synthesis and Biological Evaluation of Novel Pyrazolo[3,4-d]pyrimidine Compounds. Bioorg. Chem. 2019, 86, 393–400. [Google Scholar] [CrossRef] [PubMed]
- El-Husseiny, W.M.; El-Sayed, M.A.-A.; El-Azab, A.S.; AlSaif, N.A.; Alanazi, M.M.; Abdel-Aziz, A.A.-M. Synthesis, antitumor activity, and molecular docking study of 2-cyclopentyloxyanisole derivatives: Mechanistic study of enzyme inhibition. J. Enzyme Inhib. Med. Chem. 2020, 35, 744–758. [Google Scholar] [CrossRef]
- Redzicka, A. COX-1/COX-2 inhibition activities and molecular docking study of newly designed and synthesized pyrrolo[3,4-c]pyrrole Mannich bases. Bioorg. Med. Chem. 2019, 27, 3918–3928. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Y.; Zhang, Y.-Y.; Chu, G.-X.; Bao, G.-H. N-ethyl-2-pyrrolidinone substitution enhances binding affinity between tea flavoalkaloids and human serum albumin: Greatly influenced by esterization. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 262, 120097. [Google Scholar] [CrossRef] [PubMed]
- Rout, J.; Swain, B.C.; Mishra, P.P.; Tripathy, U. Spectroscopic insight into the interaction of dopamine with spherical gold nanoparticles. J. Photochem. Photobiol. B 2020, 203, 111770. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, Z.; Ni, Y. Interaction between aspirin and vitamin C with human serum albumin as binary and ternary systems. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 236, 118356. [Google Scholar] [CrossRef] [PubMed]
- Acharya, D.P.; Sanguansri, L.; Augustin, M.A. Binding of resveratrol with sodium caseinate in aqueous solutions. Food Chem. 2013, 141, 1050–1054. [Google Scholar] [CrossRef]
- Han, X.; Hao, H.; Li, Q.; Liu, C.; Lei, J.; Yu, F.; Chen, K.; Liu, Y.; Huang, T. The interaction mechanism between fludarabine and human serum albumin researched by comprehensive spectroscopic methods and molecular docking technique. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 233, 118170. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Y.; Chen, M.; Xie, B.; Yang, J.; Chen, J.; Sun, Z. Isorenieratene interaction with human serum albumin: Multi-spectroscopic analyses and docking simulation. Food Chem. 2018, 258, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Dai, T.; Li, T.; Huang, K.; Li, Y.; Liu, C.; Chen, J. Investigation on the binding interaction between rice glutelin and epigallocatechin-3-gallate using spectroscopic and molecular docking simulation. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 217, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, J.; Yan, J.; Wu, D.; Li, H. Spectroscopy and docking simulations of the interaction between lochnericine and bovine serum albumin: Interaction between lochnericine and bovine serum albumin. Luminescence 2015, 30, 240–246. [Google Scholar] [CrossRef]
- Seal, P.; Sikdar, J.; Ghosh, N.; Biswas, P.; Haldar, R. Exploring the binding dynamics of etoricoxib with human hemoglobin: A spectroscopic, calorimetric, and molecular modeling approach. J. Biomol. Struct. Dyn. 2019, 37, 3018–3028. [Google Scholar] [CrossRef] [PubMed]
- Jash, C.; Basu, P.; Payghan, P.V.; Ghoshal, N.; Kumar, G.S. Chelerythrine-lysozyme interaction: Spectroscopic studies, thermodynamics and molecular modeling exploration. Phys. Chem. Chem. Phys. PCCP 2015, 17, 16630–16645. [Google Scholar] [CrossRef]
- Mousavi, S.F.; Fatemi, M.H. Probing the binding mechanism of capecitabine to human serum albumin using spectrometric methods, molecular modeling, and chemometrics approach. Bioorg. Chem. 2019, 90, 103037. [Google Scholar] [CrossRef]
- Gu, J.; Yang, G.; Huang, X.; He, Q. Revealing the complexity of distinct manganese species-protein interactions through multi-spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 260, 119981. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Sun, H.; Guo, J.; Fan, J.; Li, G.; Xu, S. Molecular mechanism of the interaction between resveratrol and trypsin via spectroscopy and molecular docking. Food Funct. 2019, 10, 3291–3302. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Y.; Meng, X.; Wang, X.; Li, Z.; Qian, S.; Wei, Y.; Shu, L.; Ding, Y.; Wang, P.; et al. Total C-21 steroidal glycosides, isolated from the root tuber of Cynanchum auriculatum Royle ex Wight, attenuate hydrogen peroxide-induced oxidative injury and inflammation in L02 cells. Int. J. Mol. Med. 2018, 42, 3157–3170. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, J.-P.; Zhu, S.-X.; Guan, W.-J.; Chen, M.; Zhong, N.-S. Carbocisteine attenuates hydrogen peroxide-induced inflammatory injury in A549 cells via NF-κB and ERK1/2 MAPK pathways. Int. Immunopharmacol. 2015, 24, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Chae, H.-J.; Park, S.-Y.; Kim, J.-H. Porcine placenta hydrolysates enhance osteoblast differentiation through their antioxidant activity and effects on ER stress. BMC Complement. Altern. Med. 2016, 16, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andérica-Romero, A.C.; Hernández-Damián, J.; Vázquez-Cervantes, G.I.; Torres, I.; Pedraza-Chaverri, J. The MLN4924 inhibitor exerts a neuroprotective effect against oxidative stress injury via Nrf2 protein accumulation. Redox Biol. 2016, 8, 341–347. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.H.; Zuo, K.Y.; Zhang, Y.Y.; Wang, B.; Han, X.; Lian, A.B.; Liu, J.Y. NANOG Alleviates the Damage of Human Hair Follicle Mesenchymal Stem Cells Caused by H2O2 through Activation of AKT Pathway. Biomed. Environ. Sci. BES 2019, 32, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Chu, J.; Chen, H.; Cheng, H.; Su, J.; Wang, X.; Cao, Y.; Tian, S.; Li, Q. Gastrodin Inhibits H2O2-Induced Ferroptosis through Its Antioxidative Effect in Rat Glioma Cell Line C6. Biol. Pharm. Bull. 2020, 43, 480–487. [Google Scholar] [CrossRef] [Green Version]
- Laigre, E.; Goyard, D.; Tiertant, C.; Dejeu, J.; Renaudet, O. The study of multivalent carbohydrate–protein interactions by bio-layer interferometry †Electronic supplementary information (ESI) available. Org. Biomol. Chem. 2018, 16, 8899–8903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Liu, S.; Liu, R.; He, J. Bioassay-guided isolation of cyclooxygenase-2 inhibitory and antioxidant phenylpropanoid derivatives from the roots of Dendropanax dentiger. Bioorg. Chem. 2020, 104, 104211. [Google Scholar] [CrossRef] [PubMed]
- Doriguetto, A.C.; Martins, F.T.; Ellena, J.; Salloum, R.; dos Santos, M.H.; Moreira, M.E.C.; Schneedorf, J.M.; Nagem, T.J. 2,2’,4-trihydroxybenzophenone: Crystal structure, and anti-inflammatory and antioxidant activities. Chem. Biodivers. 2007, 4, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Lin, X.; Lu, X.; Wan, J.; Zhou, X.; Liao, S.; Tu, Z.; Xu, S.; Liu, Y. Sesquiterpenoids and xanthones derivatives produced by sponge-derived fungus Stachybotry sp. HH1 ZSDS1F1-2. J. Antibiot. 2015, 68, 121–125. [Google Scholar] [CrossRef]
- Gautam, R.; Jachak, S.M.; Kumar, V.; Mohan, C.G. Synthesis, biological evaluation and molecular docking studies of stellatin derivatives as cyclooxygenase (COX-1, COX-2) inhibitors and anti-inflammatory agents. Bioorg. Med. Chem. Lett. 2011, 21, 1612–1616. [Google Scholar] [CrossRef]
- Martins, F.T.; Assis, D.M.; dos Santos, M.H.; Camps, I.; Veloso, M.P.; Juliano, M.A.; Alves, L.C.; Doriguetto, A.C. Natural polyprenylated benzophenones inhibiting cysteine and serine proteases. Eur. J. Med. Chem. 2009, 44, 1230–1239. [Google Scholar] [CrossRef]
- Chainoglou, E.; Siskos, A.; Pontiki, E.; Hadjipavlou-Litina, D. Hybridization of Curcumin Analogues with Cinnamic Acid Derivatives as Multi-Target Agents Against Alzheimer’s Disease Targets. Molecules 2020, 25, 14958. [Google Scholar] [CrossRef]









| Compound | T (K) | Ksv (×104 L∙mol−1) | Kq (×1012 L∙mol−1∙S−1) | Ra | KA (L∙mol−1) | n | Rb |
|---|---|---|---|---|---|---|---|
| DB2 | 298 | 1.43 ± 0.188 | 1.43 ± 0.188 | 0.994 | 35,432 ± 4052.5 ** | 1.10 ± 0.0208 | 0.994 |
| 310 | 1.14 ± 0.380 | 1.14 ± 0.380 | 0.972 | 13,677 ± 2797.7 * | 1.03 ± 0.0633 | 0.986 | |
| SC2 | 298 | 2.31 ± 0.988 | 2.31 ± 0.988 | 0.991 | 23,955 ± 2224.7 ** | 1.01 ± 0.0469 | 0.996 |
| 310 | 2.07 ± 1.03 | 2.07 ± 1.03 | 0.978 | 21,881 ± 4951.0 ** | 1.02 ± 0.0731 | 0.987 | |
| YB2 | 298 | 3.49 ± 1.69 | 3.49 ± 1.69 | 0.995 | 63,827 ± 2612.1 ** | 1.07 ± 0.0551 | 0.993 |
| 310 | 2.58 ± 0.562 | 2.58 ± 0.562 | 0.995 | 46,411 ± 5519.0 ** | 1.07 ± 0.0306 | 0.995 | |
| Control compound | 298 | 1.71 ± 0.297 | 1.71 ± 0.297 | 0.990 | 4436.3 ± 718.24 | 0.947 ± 0.116 | 0.982 |
| 310 | 1.05 ± 0.157 | 1.05 ± 0.157 | 0.981 | 1417.0 ± 92.261 | 0.785 ± 0.0195 | 0.992 |
| Compound | T (K) | ΔH (kJ∙mol−1) | ΔS (J∙K−1) | ΔG (kJ∙mol−1) | J (×10−13 cm3∙L∙mol−1) | r (nm) |
|---|---|---|---|---|---|---|
| DB2 | 298 | −61.6 ± 8.20 | −120 ± 28.1 | −25.9 ± 0.280 | 1.64 | 6.42 |
| 310 | −24.6 ± 0.561 | |||||
| SC2 | 298 | −6.74 ± 9.11 | 61.2 ± 31.3 | −25.0 ± 0.229 | 1.55 | 6.62 |
| 310 | −25.7 ± 0.599 | |||||
| YB2 | 298 | −20.7 ± 6.48 | 22.6 ± 21.9 | −27.4 ± 0.101 | 1.72 | 6.47 |
| 310 | −27.7 ± 0.318 |
| System | Peak | Peak Position λex/λem (nm/nm) | Relative Intensity (I) |
|---|---|---|---|
| COX2 | 1 | 272/338 | 874 |
| 2 | 230/337 | 775 | |
| COX2-DB2 | 1 | 272/336.5 | 665 |
| 2 | 236/337 | 153 | |
| COX2-SC2 | 1 | 275/332.5 | 395 |
| 2 | 236/333 | 82.9 | |
| COX2-YB2 | 1 | 272/334.5 | 472 |
| 2 | 236/336 | 103 |
| Compound | Protective Rate (%) a,b | EC50 (nM) a,b |
|---|---|---|
| DB1 | 52.70 ± 2.021 ##, * | 7080 ± 92.56 |
| DB2 | 59.12 ± 5.571 ## | 1360 ± 463.4 |
| DC1 | 60.30 ± 7.399 ## | 3310 ± 242.5 |
| DC2 | 67.34 ±6.136 | 1550 ± 924.4 |
| SB1 | 75.83 ±7.060 | 12.01 ± 4.805 |
| SB2 | 76.91 ± 7.558 | 25.22 ± 12.16 |
| SC1 | 80.62 ± 3.605 | 1910 ± 521.3 |
| SC2 | 90.02 ± 5.410 ** | 6.254 ± 0.4153 |
| YB1 | 54.73 ± 1.900 ##, * | 8510 ± 956.8 |
| YB2 | 69.32 ± 6.164 | 417.5 ± 24.56 |
| YC1 | 55.20 ± 3.665 ## | 1910 ± 823.4 |
| YC2 | 67.01 ± 7.390 | 3850 ± 925.3 |
| Fenofibrate | 78.22 ± 9.861 | 6920 ± 610.2 |
| Quereetin | 69.53 ± 3.284 | 7980 ± 1230 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, Y.; Qin, Y.; Liu, L.; Chen, X.; Li, Y.; Li, Q. An Investigation into the Interaction between Double Hydroxide-Based Antioxidant Benzophenone Derivatives and Cyclooxygenase 2. Molecules 2021, 26, 6622. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26216622
Qiao Y, Qin Y, Liu L, Chen X, Li Y, Li Q. An Investigation into the Interaction between Double Hydroxide-Based Antioxidant Benzophenone Derivatives and Cyclooxygenase 2. Molecules. 2021; 26(21):6622. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26216622
Chicago/Turabian StyleQiao, Yanan, Yuxi Qin, Lihua Liu, Xi Chen, Yunlan Li, and Qingshan Li. 2021. "An Investigation into the Interaction between Double Hydroxide-Based Antioxidant Benzophenone Derivatives and Cyclooxygenase 2" Molecules 26, no. 21: 6622. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26216622