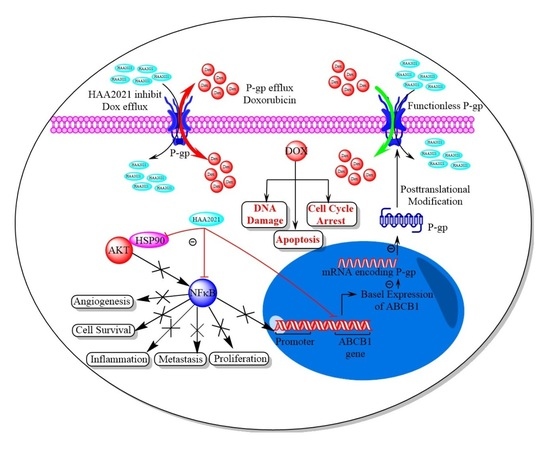
Co-Inhibition of P-gp and Hsp90 by an Isatin-Derived Compound Contributes to the Increase of the Chemosensitivity of MCF7/ADR-Resistant Cells to Doxorubicin
Abstract
:1. Introduction
2. Results
2.1. Synergistic Effects of Doxorubicin and HAA2021
2.1.1. Cytotoxicity and Selectivity of Doxorubicin and HAA2021 against Six Cell-Lines
2.1.2. Increased Chemosensitivity of MCF7/ADR Cells to Doxorubicin following Synergistic Combination with HAA2021
2.1.3. Synergistic Apoptotic Effect of Doxorubicin and HAA2021 in MCF7 and MCF7/ADR Cells
2.2. Inhibition of P-gp by HA2021
2.2.1. Molecular Modeling of HA2021/P-gp
2.2.2. Concentration-Dependent Inhibition of P-gp in MCF7/ADR Cells by HAA2021 Using Rho123 Efflux and Accumulation Assays
2.3. Inhibition of Hsp90α by HA2021
2.3.1. Surface Plasmon Resonance Analyses of HA2021/Hsp90α
2.3.2. Molecular Modeling of HA2021/Hsp90α
2.3.3. HAA2021 Inhibits Hsp90α and NF-κB in MCF7 and MCF7/ADR Cells
2.4. Synergistic Inhibition of P-gp/Hsp90α in MCF7/ADR Cells by Doxorubicin and HAA2021
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Methods
4.2.1. Cells and Maintenance
4.2.2. MTT Cytotoxicity, Selectivity, and Combination Assays
4.2.3. Determination of Apoptosis by Flowcytometery
4.2.4. Molecular Docking Calculations
Molecular Dynamic (MD) Simulations
Binding Energy Evaluation
4.2.5. Rhodamine123 Efflux Assay by Flowcytometery
4.2.6. Rhodamine123 Accumulation Assay by Spectrofluorometer
4.2.7. Surface Plasmon Resonance Analyses
4.2.8. Immunofluorescence Staining
4.2.9. Quantitative Real Time-PCR
4.2.10. Statistics and Drawing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| 17-AAG | 17-N-allylamino-17-demethoxygeldanamycin |
| ABC | ATP-binding cassette transporters |
| CI | combination index |
| EGF | epidermal growth factors |
| EGFR | epidermal growth factor receptors |
| Hsp90 | heat shock protein 90 |
| NF-𝜅B | nuclear factor kappa B |
| PI3K | phosphoinositide-3-kinase |
| P-gp | P-glycoprotein |
| Rho123 | Rhodamine-123 |
| VEGF | vascular endothelial growth factors |
| VEGFR | vascular endothelial growth factor receptors |
References
- Talevi, A. Multi-target pharmacology: Possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front Pharm. 2015, 6, 205. [Google Scholar] [CrossRef] [Green Version]
- Neophytou, C.M.; Trougakos, I.P.; Erin, N.; Papageorgis, P. Apoptosis Deregulation and the Development of Cancer Multi-Drug Resistance. Cancers 2021, 13, 4363. [Google Scholar] [CrossRef]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Li, H.; Li, Y.; Ruan, Y.; Quan, C. Identification of genes and pathways associated with MDR in MCF-7/MDR breast cancer cells by RNA-seq analysis. Mol. Med. Rep. 2018, 17, 6211–6226. [Google Scholar] [CrossRef] [Green Version]
- Abdalla, A.N.; Malki, W.H.; Qattan, A.; Shahid, I.; Hossain, M.A.; Ahmed, M. Chemosensitization of HT29 and HT29-5FU Cell Lines by a Combination of a Multi-Tyrosine Kinase Inhibitor and 5FU Downregulates ABCC1 and Inhibits PIK3CA in Light of Their Importance in Saudi Colorectal Cancer. Molecules 2021, 26, 334. [Google Scholar] [CrossRef]
- El-Azab, A.S.; Al-Dhfyan, A.; Abdel-Aziz, A.A.M.; Abou-Zeid, L.A.; Alkahtani, H.M.; Al-Obaid, A.M.; Al-Gendy, M.A. Synthesis, anticancer and apoptosis-inducing activities of quinazoline–isatin conjugates: Epidermal growth factor receptor-tyrosine kinase assay and molecular docking studies. J. Enzym. Inhib. Med. Chem. 2017, 27, 682–693. [Google Scholar] [CrossRef] [Green Version]
- Korashy, H.M.; Maayah, Z.H.; Al Anazi, F.E.; Alsaad, A.M.; Alanazi, I.O.; Belali, O.M.; Al-Atawi, F.O.; Alshamsan, A. Sunitinib inhibits breast cancer cell proliferation by inducing apoptosis, cell-cycle arrest and DNA repair while inhibiting NF-κB signaling pathways. Anticancer Res. 2017, 37, 4899–4909. [Google Scholar]
- Xia, L.; Tan, S.; Zhou, Y.; Lin, J.; Wang, H.; Oyang, L.; Tian, Y.; Liu, L.; Su, M.; Wang, H.; et al. Role of the NFκB-signaling pathway in cancer. OncoTargets Ther. 2018, 11, 2063–2073. [Google Scholar] [CrossRef] [Green Version]
- Tsou, S.H.; Chen, T.M.; Hsiao, H.T.; Chen, Y.H. A critical dose of doxorubicin is required to alter the gene expression profiles in MCF-7 cells acquiring multidrug resistance. PLoS ONE 2015, 10, e0116747. [Google Scholar] [CrossRef]
- Bruns, A.F.; Yuldasheva, N.; Latham, A.M.; Bao, L.; Pellet-Many, C.; Frankel, P.; Stephen, S.L.; Howell, G.J.; Wheatcroft, S.B.; Kearney, M.T.; et al. A heat-shock protein axis regulates VEGFR2 proteolysis, blood vessel development and repair. PLoS ONE 2012, 7, e48539. [Google Scholar] [CrossRef]
- Pandeya, S.N.; Smitha, S.; Jyoti, M.; Sridhar, S.K. Biological activities of isatin and its derivatives. Acta Pharm. 2005, 55, 27–46. [Google Scholar] [PubMed]
- Fouche, G.; Cragg, G.M.; Pillay, P.; Kolesnikova, N.; Maharaj, V.J.; Senabe, J. In vitro anticancer screening of South African plants. J. Ethnopharmacol. 2008, 119, 455–461. [Google Scholar] [CrossRef]
- Ren, X.; Xie, X.; Chen, B.; Liu, L.; Jiang, C.; Qian, Q. Marine Natural Products: A Potential Source of Anti-hepatocellular Carcinoma Drugs. J. Med. Chem. 2021, 64, 7879–7899. [Google Scholar] [CrossRef]
- Ringel, I.; Horwitz, S.B. Studies with RP 56976 (taxotere): A semisynthetic analogue of taxol. J. Natl. Cancer Inst. 1991, 83, 288–291. [Google Scholar] [CrossRef]
- Chadha, N.; Silakari, O. Indoles as therapeutics of interest in medicinal chemistry: Bird’s eye view. Eur. J. Med. Chem. 2017, 134, 159–184. [Google Scholar] [CrossRef]
- Havrylyuk, D.; Kovach, N.; Zimenkovsky, B.; Vasylenko, O.; Lesyk, R. Synthesis and anticancer activity of isatin-based pyrazolines and thiazolidines conjugates. Arch. Pharm. 2011, 344, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Singh, J.V.; Gupta, M.K.; Saxena, A.K.; Sharma, S.; Nepali, K.; Bedi, P.M.S. Triazole tethered isatin-coumarin based molecular hybrids as novel antitubulin agents: Design, synthesis, biological investigation and docking studies. Bioorg. Med. Chem. Lett. 2017, 27, 3974–3979. [Google Scholar] [CrossRef]
- Hamed, A.R.; Abdel-Azim, N.S.; Shams, K.A.; Hammouda, F.M. Targeting multidrug resistance in cancer by natural chemosensitizers. Bull. Natl. Res. Cent. 2019, 43, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chen, F.; Wang, J.; Chen, Y.; Zhang, Z.; Lin, Y.; Zhu, X. Novel isatin derivatives of podophyllotoxin: Synthesis and cytotoxic evaluation against human leukaemia cancer cells as potent anti-MDR agents. RSC Adv. 2015, 5, 97816–97823. [Google Scholar] [CrossRef]
- Rajesh Kumar, M.; Violet Dhayabaran, V.; Sudhapriya, N.; Manikandan, A.; Gideon, D.A.; Annapoorani, S. p-TSA. H2O mediated one-pot, multi-component synthesis of isatin derived imidazoles as dual-purpose drugs against inflammation and cancer. Bioorg. Chem. 2020, 102, 104046. [Google Scholar] [CrossRef]
- Havrylyuk, D.; Zimenkovsky, B.; Vasylenko, O.; Gzella, A.; Lesyk, R. Synthesis of new 4-thiazolidinone-, pyrazoline-, and isatin-based conjugates with promising antitumor activity. J. Med. Chem. 2012, 55, 8630–8641. [Google Scholar] [CrossRef]
- Medvedev, A.; Buneeva, O.; Gnedenko, O.; Ershov, P.; Ivanov, A. Isatin, an endogenous nonpeptide biofactor: A review of its molecular targets, mechanisms of actions, and their biomedical implications. BioFactors 2018, 44, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Pakravan, P.; Kashanian, S.; Khodaei, M.M.; Harding, F.J. Biochemical and pharmacological characterization of isatin and its derivatives: From structure to activity. Pharmacol. Rep. 2013, 65, 313–335. [Google Scholar] [CrossRef]
- Evdokimov, N.M.; Magedov, I.V.; McBrayer, D.; Kornienko, A. Isatin derivatives with activity against apoptosis-resistant cancer cells. Bioorg. Med. Chem. Lett. 2016, 26, 1558–1560. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.; Rothfuss, J.; Zhou, D.; MacH, R.H. Synthesis and evaluation of isatin analogs as caspase-3 inhibitors: Introduction of a hydrophilic group increases potency in a whole cell assay. Bioorg. Med. Chem. Lett. 2011, 21, 2192–2197. [Google Scholar] [CrossRef] [Green Version]
- Chinchar, E.; Makey, K.L.; Gibson, J.; Chen, F.; Cole, S.A.; Megason, G.C.; Vijayakumar, S.; Miele, L.; Gu, J.W. Sunitinib significantly suppresses the proliferation, migration, apoptosis resistance, tumor angiogenesis and growth of triple-negative breast cancers but increases breast cancer stem cells. Vasc. Cell 2014, 6, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkahtani, H.M.; Alanazi, M.M.; Aleanizy, F.S.; Alqahtani, F.Y.; Alhoshani, A.; Alanazi, F.E.; Almehizia, A.A.; Abdalla, A.N.; Alanazi, M.G.; El-Azab, A.S.; et al. Synthesis, anticancer, apoptosis-inducing activities and EGFR and VEGFR2 assay mechanistic studies of 5,5-diphenylimidazolidine-2,4-dione derivatives: Molecular docking studies. Saudi Pharm. J. 2019, 27, 682–693. [Google Scholar] [CrossRef]
- Nosol, K.; Romane, K.; Irobalieva, R.N.; Alam, A.; Kowal, J.; Fujita, N.; Locher, K.P. Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Proc. Natl. Acad. Sci. USA 2020, 117, 26245–26253. [Google Scholar] [CrossRef]
- Cooper, M.A. Label-free screening of bio-molecular interactions. Anal. Bioanal. Chem. 2003, 77, 834–842. [Google Scholar] [CrossRef]
- Caputo, M.; De Rosa, M.C.; Rescigno, T.; Zirpoli, H.; Vassallo, A.; De Tommasi, N.; Torino, G.; Tecce, M.F. Binding of polyunsaturated fatty acids to LXRα and modulation of SREBP-1 interaction with a specific SCD1 promoter element. Cell Biochem. Funct. 2014, 32, 637–646. [Google Scholar] [CrossRef]
- Malafronte, N.; Vassallo, A.; Dal Piaz, F.; Bader, A.; Braca, A.; De Tommasi, N. Biflavonoids from Daphne linearifolia Hart. Phytochem. Lett. 2012, 5, 621–625. [Google Scholar] [CrossRef]
- Dal Piaz, F.; Vera Saltos, M.B.; Franceschelli, S.; Forte, G.; Marzocco, S.; Tuccinardi, T.; Poli, G.; Nejad Ebrahimi, S.; Hamburger, M.; De Tommasi, N. Drug Affinity Responsive Target Stability (DARTS) Identifies Laurifolioside as a New Clathrin Heavy Chain Modulator. J. Nat. Prod. 2016, 79, 2681–2692. [Google Scholar] [CrossRef]
- Terracciano, S.; Chini, M.G.; Vaccaro, M.C.; Strocchia, M.; Foglia, A.; Vassallo, A.; Saturnino, C.; Riccio, R.; Bifulco, G.; Bruno, I. Identification of the key structural elements of a dihydropyrimidinone core driving toward more potent Hsp90 C-terminal inhibitors. Chem. Commun. 2016, 52, 12857–12860. [Google Scholar] [CrossRef] [PubMed]
- Schulte, T.W.; Akinaga, S.; Soga, S.; Sullivan, W.; Stensgard, B.; Toft, D.; Neckers, L.M. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 1998, 3, 100. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Reigan, P.; Siegel, D.; Zirrolli, J.; Gustafson, D.; Ross, D. Formation of 17-allylamino-demethoxygeldanamycin (17-AAG) hydroquinone by NAD(P)H:quinone oxidoreductase 1: Role of 17-AAG hydroquinone in heat shock protein 90 inhibition. Cancer Res. 2005, 65, 10006–10015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camero, C.M.; Vassallo, A.; De Leo, M.; Temraz, A.; De Tommasi, N.; Braca, A. Limonoids from Aphanamixis polystachya Leaves and Their Interaction with Hsp90. Planta Med. 2018, 84, 964–970. [Google Scholar] [CrossRef] [Green Version]
- Amaral, M.; Kokh, D.B.; Bomke, J.; Wegener, A.; Buchstaller, H.P.; Eggenweiler, H.M.; Matias, P.; Sirrenberg, C.; Wade, R.C.; Frech, M. Protein conformational flexibility modulates kinetics and thermodynamics of drug binding. Nat. Commun. 2017, 8, 2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poli, G.; Lapillo, M.; Granchi, C.; Caciolla, J.; Mouawad, N.; Caligiuri, I.; Rizzolio, F.; Langer, T.; Minutolo, F.; Tuccinardi, T. Binding investigation and preliminary optimisation of the 3-amino-1,2,4-triazin-5(2H)-one core for the development of new Fyn inhibitors. J. Enzym. Inhib. Med. Chem. 2018, 33, 956–961. [Google Scholar] [CrossRef] [Green Version]
- Dal Piaz, F.; Ferro, P.; Vassallo, A.; Vasaturo, M.; Forte, G.; Chini, M.G.; Bifulco, G.; Tosco, A.; De Tommasi, N. Identification and mechanism of action analysis of the new PARP-1 inhibitor 2″-hydroxygenkwanol A. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1806–1814. [Google Scholar] [CrossRef]
- Domotor, O.; Tuccinardi, T.; Karcz, D.; Walsh, M.; Creaven, B.S.; Enyedy, E.A. Interaction of anticancer reduced Schiff base coumarin derivatives with human serum albumin investigated by fluorescence quenching and molecular modeling. Bioorg. Chem. 2014, 52, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Milella, L.; Milazzo, S.; De Leo, M.; Vera Saltos, M.B.; Faraone, I.; Tuccinardi, T.; Lapillo, M.; De Tommasi, N.; Braca, A. Alpha-Glucosidase and alpha-Amylase Inhibitors from Arcytophyllum thymifolium. J. Nat. Prod. 2016, 79, 2104–2112. [Google Scholar] [CrossRef]
- Granchi, C.; Caligiuri, I.; Bertelli, E.; Poli, G.; Rizzolio, F.; Macchia, M.; Martinelli, A.; Minutolo, F.; Tuccinardi, T. Development of terphenyl-2-methyloxazol-5(4H)-one derivatives as selective reversible MAGL inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 1240–1252. [Google Scholar] [CrossRef] [Green Version]
- Vine, K.L.; Belfiore, L.; Jones, L.; Locke, J.M.; Wade, S.; Minaei, E.; Ranson, M. N-alkylated isatins evade P-gp mediated efflux and retain potency in MDR cancer cell lines. Heliyon 2016, 2, e00060. [Google Scholar] [CrossRef] [Green Version]
- Sevin, M.; Girodon, F.; Garrido, C.; de Thonel, A. HSP90 and HSP70: Implication in Inflammation Processes and Therapeutic Approaches for Myeloproliferative Neoplasms. Mediat. Inflamm. 2015, 2015, 970242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentires-Alj, M.; Barbu, V.; Fillet, M.; Chariot, A.; Relic, B.; Jacobs, N.; Gielen, J.; Merville, M.P.; Bours, V. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 2003, 22, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.B.; Lee, S.H.; Um, J.H.; Oh, W.K.; Kim, D.W.; Kang, C.D.; Kim, S.H. Sensitization of multidrug-resistant human cancer cells to Hsp90 inhibitors by down-regulation of SIRT1. Oncotarget 2015, 6, 36202–36218. [Google Scholar] [CrossRef]
- Yin, L.; Yang, Y.; Zhu, W.; Xian, Y.; Han, Z.; Huang, H.; Peng, L.; Zhang, K.; Zhao, Y. Heat Shock Protein 90 Triggers Multi-Drug Resistance of Ovarian Cancer via AKT/GSK3beta/beta-Catenin Signaling. Front. Oncol 2021, 11, 620907. [Google Scholar] [CrossRef]
- Dinic, J.; Podolski-Renic, A.; Jovanovic, M.; Musso, L.; Tsakovska, I.; Pajeva, I.; Dallavalle, S.; Pesic, M. Novel Heat Shock Protein 90 Inhibitors Suppress P-Glycoprotein Activity and Overcome Multidrug Resistance in Cancer Cells. Int. J. Mol. Sci. 2019, 20, 4575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bader, A.; Bkhaitan, M.M.; Abdalla, A.N.; Abdallah, Q.M.A.; Ali, H.I.; Sabbah, D.A.; Albadawi, G.; Abushaikha, G.M. Design and Synthesis of 4-O-Podophyllotoxin Sulfamate Derivatives as Potential Cytotoxic Agents. Evid. Based Complement. Altern. Med. 2021, 2021, 6672807. [Google Scholar] [CrossRef] [PubMed]
- Al-Salem, H.S.; Arifuzzaman, M.; Alkahtani, H.M.; Abdaalla, A.N.; Issa, I.S.; Alqathama, A.; Albalawi, F.S.; Rahman, A. A Series of Isatin-Hydrazones with Cytotoxic Activity and CDK2 Kinase Inhibitory Activity: A Potential Type II ATP Competitive Inhibitor. Molecules 2020, 25, 4400. [Google Scholar] [CrossRef]
- Al Bratty, M.; Makeen, H.A.; Alhazmi, H.A.; Syame, S.M.; Abdalla, A.N.; Homeida, H.E.; Sultana, S.; Ahsan, W.; Khalid, A. Phytochemical, Cytotoxic, and Antimicrobial Evaluation of the Fruits of Miswak Plant, Salvadora persica L. J. Chem. 2020, 2020, 452195. [Google Scholar] [CrossRef]
- Abdalla, A.N.; Abdallah, M.E.; Aslam, A.; Bader, A.; Vassallo, A.; Tommasi, N.; Malki, W.H.; Gouda, A.M.; Mukhtar, M.H.; El-Readi, M.Z.; et al. Synergistic Anti Leukemia Effect of a Novel Hsp90 and a Pan Cyclin Dependent Kinase Inhibitors. Molecules 2020, 25, 2220. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, A.N.; Shaheen, U.; Abdallah, Q.M.A.; Flamini, G.; Bkhaitan, M.M.; Abdelhady, M.I.S.; Ascrizzi, R.; Bader, A. Proapoptotic Activity of Achillea membranacea Essential Oil and Its Major Constituent 1,8-Cineole against A2780 Ovarian Cancer Cells. Molecules 2020, 25, 1582. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Granchi, C.; Lapillo, M.; Glasmacher, S.; Bononi, G.; Licari, C.; Poli, G.; El Boustani, M.; Caligiuri, I.; Rizzolio, F.; Gertsch, J.; et al. Optimization of a Benzoylpiperidine Class Identifies a Highly Potent and Selective Reversible Monoacylglycerol Lipase (MAGL) Inhibitor. J. Med. Chem. 2019, 62, 1932–1958. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Poli, G.; Gelain, A.; Porta, F.; Asai, A.; Martinelli, A.; Tuccinardi, T. Identification of a new STAT3 dimerization inhibitor through a pharmacophore-based virtual screening approach. J. Enzym. Inhib. Med. Chem. 2016, 31, 1011–1017. [Google Scholar] [CrossRef] [Green Version]
- Eid, S.Y.; Althubiti, M.A.; Abdallah, M.E.; Wink, M.; El-Readi, M.Z. The carotenoid fucoxanthin can sensitize multidrug resistant cancer cells to doxorubicin via induction of apoptosis, inhibition of multidrug resistance proteins and metabolic enzymes. Phytomedicine 2020, 77, 153280. [Google Scholar] [CrossRef] [PubMed]
- Eid, S.Y.; El-Readi, M.Z.; Eldin, E.E.; Fatani, S.H.; Wink, M. Influence of combinations of digitonin with selected phenolics, terpenoids, and alkaloids on the expression and activity of P-glycoprotein in leukaemia and colon cancer cells. Phytomedicine 2013, 21, 47–61. [Google Scholar] [CrossRef]
- Abdalla, A.N.; Qattan, A.; Malki, W.H.; Shahid, I.; Hossain, M.A.; Ahmed, M. Significance of Targeting VEGFR-2 and Cyclin D1 in Luminal-A Breast Cancer. Molecules 2020, 25, 4606. [Google Scholar] [CrossRef]
- Almasmoum, H.; Refaat, B.; Ghaith, M.M.; Almaimani, R.A.; Idris, S.; Ahmad, J.; Abdelghany, A.H.; BaSalamah, M.A.; El-Boshy, M. Protective effect of Vitamin D3 against lead induced hepatotoxicity, oxidative stress, immunosuppressive and calcium homeostasis disorders in rat. Environ. Toxicol. Pharm. 2019, 72, 103246. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.E.; El-Readi, M.Z.; Althubiti, M.A.; Almaimani, R.A.; Ismail, A.M.; Idris, S.; Refaat, B.; Almalki, W.H.; Babakr, A.T.; Mukhtar, M.H.; et al. Tamoxifen and the PI3K Inhibitor: LY294002 Synergistically Induce Apoptosis and Cell Cycle Arrest in Breast Cancer MCF-7 Cells. Molecules 2020, 25, 3355. [Google Scholar] [CrossRef] [PubMed]















| Cell Line | Doxorubicin | HAA2021 | ||
|---|---|---|---|---|
| IC50 | SI a | IC50 | SI b | |
| MCF7 | 0.05 ± 0.00 | 10.00 | 0.22 ± 0.05 | 86.59 |
| MCF7/ADR | 13.99 ± 2.10 | 0.03 | 17.21 ± 2.33 | 1.10 |
| HL60 | 0.30 ± 0.04 | 1.66 | 2.36 ± 0.69 | 8.07 |
| K562 | 0.03 ± 0.00 | 16.66 | 16.04 ± 4.45 | 1.19 |
| HT29 | 0.04 ± 0.00 | 12.50 | 5.06 ± 1.61 | 3.76 |
| MRC5 | 0.50 ± 0.09 | - | 19.05 ± 1.03 | - |
| Drug (µM) | IC50 | CI a | R b | |
|---|---|---|---|---|
| Doxorubicin | HAA2021 | |||
| - | 0–50 | 17.21 ± 2.33 | - | - |
| 0–50 | - | 13.99 ± 2.10 | - | - |
| 0.25 | 0.25 | 15.09 ± 1.03 | >100 | 0.97 |
| 0.50 | 0.25 | 14.34 ± 3.00 | 4.555 | 0.91 |
| 1.00 | 0.25 | 6.04 ± 1.01 | 0.014 | 0.97 |
| 0.25 | 0.50 | 14.64 ± 2.60 | >100 | 0.89 |
| 0.50 | 0.50 | 5.54 ± 0.87 | 0.692 | 0.93 |
| 1.00 | 0.50 | 1.01 ± 0.12 | 0.003 | 0.90 |
| 0.25 | 1.00 | 14.09 ± 2.87 | 90.244 | 0.95 |
| 0.50 | 1.00 | 13.02 ± 1.99 | 3.670 | 0.93 |
| 1.00 | 1.00 | 0.89 ± 0.07 | 0.001 | 0.94 |
| Compound | KD (nM) a |
|---|---|
| HAA2021 | 41.20 ± 2.10 |
| Radicicol | 1.80 ± 0.40 |
| 17-AAG | 360.00 ± 21.90 |
| MM−GBSA Method | |||||
| Hsp90−HAA2021 Complex | VDW | ELE | EGB | ESURF | ΔGBSA |
| Hel. CL1 | −51.9 | −8.3 | 33.8 | −6.7 | −33.1 |
| Hel. CL2 | −47.2 | −33.0 | 49.3 | −6.0 | −36.9 |
| Hel. CL3 | −45.4 | −26.1 | 44.6 | −6.2 | −33.2 |
| Hel. CL4 | −50.3 | −8.8 | 34.1 | −6.2 | −31.1 |
| Hel. CL5 | −45.4 | −10.1 | 33.3 | −5.8 | −28.1 |
| L.−in CL1 | −51.4 | −40.7 | 51.3 | −6.2 | −47.0 |
| L.−in CL2 | −40.1 | −41.2 | 56.3 | −5.5 | −30.4 |
| L.−in CL3 | −45.4 | −47.9 | 69.0 | −6.2 | −30.5 |
| L.−in CL4 | −40.5 | −18.2 | 35.7 | −5.2 | −28.1 |
| L.−out CL1 | −34.6 | −18.7 | 36.2 | −4.4 | −21.6 |
| L.−out CL2 | −41.1 | −1.6 | 23.9 | −5.2 | −23.9 |
| MM−PBSA method | |||||
| Hsp90−HAA2021 Complex | VDW | ELE | EPB | ENPOLAR | ΔPBSA |
| Hel. CL1 | −51.9 | −8.3 | 43.1 | −5.1 | −22.2 |
| Hel. CL2 | −47.2 | −33.0 | 59.0 | −4.8 | −26.0 |
| Hel. CL3 | −45.4 | −26.1 | 57.0 | −4.9 | −19.5 |
| Hel. CL4 | −50.3 | −8.8 | 43.3 | −4.8 | −20.5 |
| Hel. CL5 | −45.4 | −10.1 | 37.2 | −4.8 | −23.1 |
| L.−in CL1 | −51.4 | −40.7 | 58.9 | −4.6 | −37.8 |
| L.−in CL2 | −40.1 | −41.2 | 69.6 | −4.4 | −16.1 |
| L.−in CL3 | −45.4 | −47.9 | 76.7 | −4.4 | −21.1 |
| L.−in CL4 | −40.5 | −18.2 | 45.7 | −4.0 | −17.0 |
| L.−out CL1 | −34.6 | −18.7 | 43.8 | −3.9 | −13.5 |
| L.−out CL2 | −41.1 | −1.6 | 31.6 | −4.1 | −15.2 |
| Gene | Sequence |
|---|---|
| GAPDH | F: AGGTCGGTGTGAACGGATTTG R: TGTAGACCATGTAGTTGAGGTCA |
| P-gp | F: TGCTCAGACAGGATGTGAGTTG R: AATTACAGCAAGCCTGGAACC |
| Hsp90α | F: TTGGTTACTTCCCCGTGCTG R: GCCTTTTGCCGTAGGGTTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdalla, A.N.; Di Stefano, M.; Poli, G.; Tuccinardi, T.; Bader, A.; Vassallo, A.; Abdallah, M.E.; El-Readi, M.Z.; Refaat, B.; Algarni, A.S.; et al. Co-Inhibition of P-gp and Hsp90 by an Isatin-Derived Compound Contributes to the Increase of the Chemosensitivity of MCF7/ADR-Resistant Cells to Doxorubicin. Molecules 2022, 27, 90. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27010090
Abdalla AN, Di Stefano M, Poli G, Tuccinardi T, Bader A, Vassallo A, Abdallah ME, El-Readi MZ, Refaat B, Algarni AS, et al. Co-Inhibition of P-gp and Hsp90 by an Isatin-Derived Compound Contributes to the Increase of the Chemosensitivity of MCF7/ADR-Resistant Cells to Doxorubicin. Molecules. 2022; 27(1):90. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27010090
Chicago/Turabian StyleAbdalla, Ashraf N., Miriana Di Stefano, Giulio Poli, Tiziano Tuccinardi, Ammar Bader, Antonio Vassallo, Mohamed E. Abdallah, Mahmoud Zaki El-Readi, Bassem Refaat, Alanood S. Algarni, and et al. 2022. "Co-Inhibition of P-gp and Hsp90 by an Isatin-Derived Compound Contributes to the Increase of the Chemosensitivity of MCF7/ADR-Resistant Cells to Doxorubicin" Molecules 27, no. 1: 90. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27010090