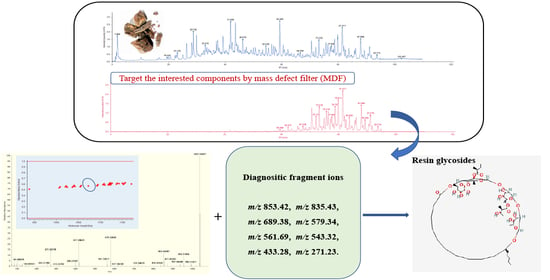
Mass Defect Filtering-Oriented Identification of Resin Glycosides from Root of Convolvulus scammonia Based on Quadrupole-Orbitrap Mass Spectrometer
Abstract
:1. Introduction
2. Results and Discussion
2.1. DFIs Determinations and Fragmentation Patterns Analysis
2.2. Construction of MDF Model
2.3. Identified Glycosidic Acids
2.4. Identified Resin Glycosides
- adduct ion [M + HCOO]−, [M + Cl]− combined with [M − H]− were observed, it is different from glycosidic acids;
- loss of short organic acid such as 2-methylbutyric acid, tiglic acid, isobutyric acid, (2R,3R)-3-hydroxy-2-methylbutyric, 3-hydroxy-2-methylenebutyric were common characteristics;
- breakage of glycosidic linkage is prone to loss of 162 Da, 146 Da;
- loss of C2H4O (44 Da) was observed when (2R,3R)-3-hydroxy-2-methylbutyric substituted on resin glycosides
- parent drug filter ion at m/z 853 with lower intensity was obtained and identical ions at m/z 579, m/z 561, m/z 543, m/z 399, m/z 417 combined with m/z 271 were discovered.
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. References
3.3. Sample Preparation
3.4. HPLC Condition
3.5. Mass Spectrometric Conditions
3.6. Mass Defect Filter Approach
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pereda-Miranda, R.; Rosas-Ramírez, D.; Castañeda-Gómez, J. Resin glycosides from the morning glory family. Prog. Chem. Org. Nat. Prod. 2010, 92, 77–147. [Google Scholar]
- Fan, B.; Lu, Y.; Yang, M.; Li, J.; Chen, G. Evolvulins I and II, resin glycosides with a trihydroxy aglycone unit from Evolvulus alsinoides. Org. Lett. 2019, 21, 6548–6551. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Takigawa, A.; Kanemaru, Y.; Kawakami, G.; Kabata, K.; Okawa, M.; Kinjo, J.; Yokomizo, K.; Yoshimitsu, H.; Nohara, T. Calysolins V–IX, resin glycosides from Calystegia soldanella and their antiviral activity toward herpes. Chem. Pharm. Bull. 2014, 62, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Ono, M. Resin glycosides from Convolvulaceae plants. J. Nat. Med. 2017, 71, 591–604. [Google Scholar] [CrossRef]
- Ono, M.; Kanemaru, Y.; Yasuda, S.; Okawa, M.; Kinjo, J.; Miyashita, H.; Yokomizo, K.; Yoshimitsu, H.; Nohara, T. A new resin glycoside from Calystegia soldanella and its antiviral activity towards herpes. Nat. Prod. Res. 2017, 31, 2660–2664. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Yagi, C.; Hama, H.; Tanaka, M.; Arihara, S.; Hashimoto, T. Ipomotaosides A–D, resin glycosides from the aerial parts of Ipomoea batatas and their inhibitory activity against COX-1 and COX-2. J. Nat. Prod. 2016, 73, 1763–1766. [Google Scholar] [CrossRef]
- Figueroa-González, G.; Jacobo-Herrera, N.; Zentella-Dehesa, A.; Pereda-Miranda, R. Reversal of multidrug resistance by morning glory resin glycosides in human breast cancer cells. J. Nat. Prod. 2012, 75, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Corona-Castaneda, B.; Rosas-Ramírez, D.; Castañeda-Gómez, J.; Aparicio-Cuevas, M.; Fragoso-Serrano, M.; Figueroa-Gonzalez, G.; Pereda-Miranda, R. Resin glycosides from Ipomoea wolcottiana as modulators of the multidrug resistance phenotype in vitro. Phytochemistry 2016, 123, 48–57. [Google Scholar] [CrossRef]
- Eich, E. Solanaceae and Convolvulaceae: Secondary Metabolites; Springer: Berlin/Heidelberg, Germany, 2008; pp. 525–582. [Google Scholar]
- Naoki, N.; Hirokuni, K.; Toshio, K.; Kazumoto, M. Scammonins I and II, the resin glycosides of radix scammoniae from Convolvulus Scammonia. Phytochemistry 1990, 29, 3565–3569. [Google Scholar]
- Hirokuni, K.; Naoki, N.; Toshio, K.; Kazumoto, M. Scammonin III–VI, resin glycosides of Convolvulus Scammonia. Phytochemistry 1991, 30, 957–963. [Google Scholar]
- Naoki, N.; Hirokuni, K.; Toshio, K.; Kazumoto, M. Scammonins VII and VIII, two resin glycosides from Convolvulus Scammonia. Phytochemistry 1992, 31, 2761–2766. [Google Scholar]
- Jia, S.L.; Du, Z.F.; Song, C.W.; Jin, S.N.; Zhang, Y.; Feng, Y.L.; Xiong, C.M.; Jiang, H.L. Identification and characterization of curcuminoids in turmeric using ultra-high performance liquid chromatography-quadrupole time of flight tandem mass spectrometry. J. Chromatogr. A 2017, 1521, 110–122. [Google Scholar] [CrossRef]
- Hao, Y.M.; Huo, J.H.; Wang, T.; Sun, G.D.; Wang, W.M. Chemical profiling of Coptis rootlet and screening of its bioactive compounds in inhibiting Staphylococcus aureus by UPLC-Q-TOF/MS. J. Pharm. Biomed. Anal. 2020, 180, 113089. [Google Scholar] [CrossRef]
- Ahad, H.; Jin, H.L.; Liu, Y.F.; Wang, J.X.; Sun, G.Y.; Liang, X.M.; Aisa, H.A. Chemical profiling of spermidines in goji berry by strong cation exchange solid-phase extraction (SCX-SPE) combined with ultrahigh-performance liq- uid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS/MS). J. Chromatogr. B 2020, 1137, 121923. [Google Scholar] [CrossRef]
- Liu, Z.H.; Wang, Y.Q.; Mei, X.D.; Wang, F.; Yang, X.Z.; Li, X.D.; Jiang, F.; Zhang, J.Y. Comprehensive analysis of the chemical constituents in sulfur-fu-migated Lonicerae Japonicae Flos using UHPLC-LTQ-Orbitrap mass spectrometry. Chin. J. Nat. Med. 2020, 18, 148–160. [Google Scholar] [PubMed]
- Cai, W.; Li, K.L.; Xiong, P.; Gong, K.Y.; Zhu, L.; Yang, J.B.; Wu, W.H. A systematic strategy for rapid identification of chlorogenic acids derivatives in Duhaldea nervosa using UHPLC-Q-Exactive Orbitrap mass spectrometry. Arab. J. Chem. 2020, 13, 3751–3761. [Google Scholar] [CrossRef]
- Qiao, S.; Shi, X.W.; Shi, R.; Liu, M.; Liu, T.; Zhang, K.R.; Wang, Q.; Yao, M.C.; Zhang, L.T. Identification of urinary metabolites of imperatorin with a single run on an LC/Triple TOF system based on multiple mass defect filter data acquisition and multiple data mining techniques. Anal. Bioanal. Chem. 2013, 405, 6721–6738. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.S.; Ma, L.; Zhang, H.Y.; Humphreys, W.G. Detection and structural characterization of glutathione-trapped reactive metabolites using liquid chromatography-high-resolution mass Spectrometry and mass defect filtering. Anal. Chem. 2007, 79, 8333–8341. [Google Scholar] [CrossRef]
- Liu, M.Y.; Zhao, S.H.; Wang, Z.Q.; Wang, Y.F.; Liu, T.; Li, S.; Wang, C.C.; Wang, H.T.; Tu, P.F. Identification of metabolites of deoxyschizandrin in rats by UPLC-Q-TOF-MS/MS based on multiple mass defect filter data acquisition and multiple data processing techniques. J. Chromatogr. B 2014, 949, 115–126. [Google Scholar] [CrossRef] [PubMed]
- An, Y.L.; Wei, W.L.; Li, H.J.; Li, Z.W.; Yao, C.L.; Qu, H.; Yao, S.; Huang, Y.; Zhang, J.Q.; Bi, Q.R.; et al. An enhanced strategy integrating offline superimposed two-dimensional separation with mass defect filter and diagnostic ion filter: Comprehensive characterization of steroid alkaloids in Fritillariae Pallidiflorae Bulbus as a case study. J. Chromatogr. A 2021, 1643, 462029. [Google Scholar] [CrossRef]
- Ding, W.B.; Jiang, Z.H.; Wu, P.; Xu, L.X.; Wei, X.Y. Resin glycosides from the aerial parts of Operculina turpethum. Phytochemistry 2012, 81, 165–174. [Google Scholar] [CrossRef]
- Xie, T.; Liang, Y.; Hao, H.J.; Xie, A.L.; Gong, P.; Dai, C.; Liu, L.; Kang, A.; Zheng, X.; Wang, G. Rapid identification of ophiopogonins and ophiopogonones in Ophiopogon japonicus extract with a practical technique of mass defect filtering based on high resolution mass spectrometry. J. Chromatogr. A 2012, 1227, 234–244. [Google Scholar] [CrossRef]
- Mortishire-Smith, R.J.; Castro-Perez, J.M.; Yu, K.; Shockcor, J.P.; Goshawk, J.; Hartshorn, M.J.; Hill, A. Generic dealkylation: A tool for increasing the hit-rate of metabolite rationalization, and automatic customization of mass defect filters. Rapid Commun. Mass Spectrom. 2009, 23, 939–948. [Google Scholar] [CrossRef]
- Claesen, J.; Dittwald, P.; Burzykowski, T.; Valkenborg, D. An efficient method to calculate the aggregated isotopic distribution and exact center-masses. J. Am. Soc. Mass Spectrom. 2012, 23, 753–763. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, D.; Ray, K.; Zhu, M. Mass defect filter technique and its applications to drug metabolite identification by high-resolution mass spectrometry. J. Mass Spectrom. 2009, 44, 999–1016. [Google Scholar] [CrossRef] [PubMed]




| References | Precursor Ion [M − H]− | Adduct Ions [M + Cl]−, [M + HCOO]− | Diagnostic Fragment Ions (m/z) | Neutral Loss (Da) |
|---|---|---|---|---|
| Turpethoside B | m/z 1265.65344 | [M + Cl]− m/z 1301.62927 | m/z 853.42, m/z 835.43, | HCOOH: 46.01 |
| m/z 689.38, m/z 579.34, | H2O: 18.01 | |||
| [M + HCOO]− m/z 1311.65771 | m/z 561.69, m/z 543.32, | Tiglic acid: 100.05 | ||
| m/z 433.28, m/z 271.23. | 2-Methylbutyric acid: 102.08 | |||
| Tupethic acid C | m/z 1047.52417 | – | m/z 593.35, m/z 574.45, | Rha: 146.06 |
| m/z 447.30, m/z 429.29, | Glc: 162.05, H2O: 18.01 | |||
| m/z 285.24, m/z 267.46. |
| Substitutes | Mass Change, Da | Mass Defect Shift, mDa |
|---|---|---|
| +2-methylbutyric acid | +C5H8O, 84.0569 | 56.9 |
| +tiglic acid | +C5H6O, 82.0413 | 41.3 |
| +isobutyric acid | +C4H6O, 70.0413 | 41.3 |
| +3-hydroxy-2-methylenebutyric | +C4H4O, 68.0257 | 25.7 |
| +3-hydroxy-2-methylbutyric | +C5H8O2, 100.0519 | 51.9 |
| +Methyl | +CH3, 14.01565 | 15.7 |
| Filters | Mass Change, Da | Mass Defect Shift, mDa | |
|---|---|---|---|
| m/z 853, C40H69O19 | +isobutyric acid | C44H75O20 (923) | 0.485 |
| +tiglic acid | C45H75O20 (935) | 0.485 | |
| +2-methylbutyric acid | C45H77O19 (937) | 0.500 | |
| +isobutyric acid, tiglic acid | C49H81O21 (1005) | 0.526 | |
| +tiglic acid, tiglic acid | C50H81O21 (1017) | 0.526 | |
| +2-methylbutyric acid, tiglic acid. | C50H83O21 (1019) | 0.542 | |
| m/z 869, C40H69O20 | +2-methylbutyric acid, tiglic acid | C50H83O22 (1035) | 0.537 |
| m/z 871, C40H71O20 | +CH3, 2-methylbutyric acid, tiglic acid | C51H87O22 (1051) | 0.568 |
| No. | Rt min | Formula | ΔMass ppm | Parent Compounds m/z | Transformations | Composition Change | MSn |
|---|---|---|---|---|---|---|---|
| 1 | 58.424 | C50H86O23 | 1.62 | 871 | 3-hydroxy-2-methylbutyric | +C10H14O3 | MS [M − H]− 1053.55042 |
| Tiglic acid | MS2 971, 953, 935, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 2 | 59.374 | C50H88O23 | 0.66 | 871 | 2-methylbutyric acid | +C10H16O3 | MS [M − H]− 1055.56519 |
| 3-hydroxy-2-methylbutyric acid | MS2 971, 955, 953, 937,871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3 | 59.504 | C55H94O26 | –0.41 | 887 | 3-hydroxy-2-methylbutyric | +C15H22O6 | MS [M − H]− 1169.59558 |
| 3-hydroxy-2-methylbutyric | MS2 969, 951, 887, 851,579, 561, 417, 399, 271 | ||||||
| Tiglic acid | |||||||
| 4 | 60.543 | C50H88O24 | 0.84 | 871 | 3-hydroxy-2-methylbutyric acid | +C10H16O4 | MS [M − H]− 1071.55969 |
| 3-hydroxy-2-methylbutyric acid | MS2 953, 909, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 5 | 62.294 | C50H86O23 | 0.69 | 871 | 3-hydroxy-2-methylbutyric | +C10H14O3 | MS [M − H]− 1053.54944 |
| Tiglic acid | MS2 971,953, 935, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 6 | 63.494 | C50H88O23 | 1.13 | 871 | 2-methylbutyric acid | +C10H16O3 | MS [M − H]− 1055.56555 |
| 3-hydroxy-2-methylbutyric acid | MS2 971, 955, 953, 937,871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 7 | 65.234 | C49H86O23 | 1.75 | 871 | isobutyric acid | +C9H14O3 | MS [M − H]− 1041.05454 |
| 3-hydroxy-2-methylbutyric acid | MS2 941, 923, 871, 853, 835, 725, 707,579, 561, 417, 399, 271 | ||||||
| 8 | 65.571 | C50H86O23 | 1.04 | 871 | 3-hydroxy-2-methylbutyric | +C10H14O3 | MS [M − H]− 1053.54980 |
| Tiglic acid | MS2 971, 953, 935, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 9 | 65.721 | C55H94O25 | 0.58 | 871 | Tiglic acid | +C15H22O5 | MS [M − H]− 1153.60181 |
| 3-hydroxy-2-methylbutyric acid | MS2 1053,953, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 10 | 65.966 | C55H94O25 | 0.68 | 871 | Tiglic acid | +C15H22O5 | MS [M − H]− 1153.60205 |
| 3-hydroxy-2-methylbutyric acid | MS2 1053, 953, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 11 | 67.468 | C55H94O25 | 0.47 | 871 | Tiglic acid | +C15H22O5 | MS [M − H]− 1153.60168 |
| 3-hydroxy-2-methylbutyric acid | MS2 1053, 953, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 12 | 68.081 | C55H94O25 | 1.10 | 871 | Tiglic acid | +C15H22O5 | MS [M − H]− 1153.60242 |
| 3-hydroxy-2-methylbutyric acid | MS2 1053, 953, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 13 | 70.587 | C55H94O25 | 0.58 | 871 | Tiglic acid | +C15H22O5 | MS [M − H]− 1153.60181 |
| 3-hydroxy-2-methylbutyric acid | MS2 1053, 953, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 14 | 71.012 | C54H92O24 | 1.30 | 871 | Tiglic acid | +C14H20O4 | MS [M − H]− 1123.59126 |
| Isobutyric acid | MS2 1023, 941, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 15 | 71.260 | C49H84O22 | 1.31 | 871 | Tiglic acid | +C9H12O2 | MS [M − H]− 1023.53931 |
| Isobutyric acid | MS2 941, 871, 853, 725, 579, 561, 417, 399, 271 | ||||||
| 16 | 71.331 | C54H92O24 | 0.98 | 871 | Tiglic acid | +C14H20O4 | MS [M − H]− 1123.59167 |
| Isobutyric acid | MS2 1023, 941, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 17 | 74.490 | C55H94O24 | 0.94 | 871 | Tiglic acid | +C15H22O4 | MS [M − H]− 1137.60730 |
| 2-methylbutyric acid | MS2 1055, 955, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 18 | 74.561 | C50H86O22 | 0.04 | 871 | 2-methylbutyric acid | +C10H14O2 | MS [M − H]− 1037.55518 |
| Tiglic acid | MS2 955, 937, 935, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 19 | 76.437 | C54H92O24 | 0.20 | 871 | Tiglic acid | +C14H20O4 | MS [M − H]− 1123.59082 |
| Isobutyric acid | MS2 1023, 941, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 20 | 76.632 | C50H88O22 | 1.42 | 871 | 2-methylbutyric acid | +C10H16O2 | MS [M − H]− 1039.57056 |
| 2-methylbutyric acid | MS2 955, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 21 | 78.369 | C54H94O24 | 0.95 | 871 | isobutyric acid | +C14H22O4 | MS [M − H]− 1125.60742 |
| 2-methylbutyric acid | MS2 1025, 941, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid | |||||||
| 22 | 79.754 | C55H94O24 | 0.84 | 871 | Tiglic acid | +C15H22O4 | MS [M − H]− 1137.60718 |
| 2-methylbutyric acid | MS2 1055, 955, 871, 853, 835, 725, 707, 579, 561, 417, 399, 271 | ||||||
| 3-hydroxy-2-methylbutyric acid |
| No. | Rt min | Formula | ΔMass ppm | Parent Compounds m/z | Transformations | Composition Change | MSn m/z |
|---|---|---|---|---|---|---|---|
| 1′ | 67.577 | C45H78O20 | 1.51 | 853 | 2-methylbutyric acid | +C5H8O | MS [M + FA − H]− 983.50909 |
| [M − H]− 937.50262 | |||||||
| MS2 853, 835, 579, 561, 417, 399, 271 | |||||||
| 2′ | 68.468 | C50H84O22 | 1.17 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54431 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.54016 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 3′ | 70.902 | C49H84O22 | 1.24 | 853 | Isobutyric acid | +C9H14O3 | MS [M + FA − H]− 1069.54492 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1023.53955 | ||||||
| MS2 935, 923, 835, 773, 671, 663, 579, 561, 417, 399, 271 | |||||||
| 4′ | 71.865 | C54H92O24 | 1.52 | 853 | Isobutyric acid | +C14H22O5 | MS [M + FA − H]− 1169.59766 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1123.59155 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1079, 1035, 1023, 935, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 5′ | 72.129 | C50H84O22 | 0.65 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54419 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.53955 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 6′ | 73.185 | C50H84O22 | 0.67 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54431 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.53943 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 7′ | 73.657 | C50H86O22 | 0.66 | 853 | 2-methylbutyric acid | +C10H16O3 | MS [M + FA − H]− 1083.55994 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1037.55225 | ||||||
| MS2 1019, 993, 937, 853, 835, 707, 663, 579, 561, 417, 399, 271 | |||||||
| 8′ | 73.905 | C50H84O22 | 1.13 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54480 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.54114 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 9′ | 74.610 | C50H86O23 | -0.31 | 853 | 3-hydroxy-2-methylbutyric acid | +C10H16O4 | MS [M + FA − H]− 1099.55291 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1053.54980 | ||||||
| MS2 1009, 909, 891, 853, 835, 737, 661, 579, 561, 417, 399, 271 | |||||||
| 10′ | 74.845 | C50H86O22 | -0.61 | 853 | 2-methylbutyric acid | +C10H16O3 | MS [M + FA − H]− 1083.55847 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1037.55383 | ||||||
| MS2 1019, 993, 935, 853, 835, 707, 663, 579, 561, 417, 399, 271 | |||||||
| 11′ | 74.977 | C50H84O22 | 0.76 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54431 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.53748 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 579, 561, 417, 399, 271 | |||||||
| 12′ | 75.092 | C55H92O24 | 0.65 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59656 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59160 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 13′ | 75.702 | C50H84O22 | 0.50 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54419 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.53918 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 14′ | 76.552 | C50H86O23 | 0.87 | 853 | 3-hydroxy-2-methylbutyric acid | +C10H16O4 | MS [M + FA − H]− 1099.55505 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1053.54810 | ||||||
| MS2 1009, 909, 891, 853, 835, 737, 661, 579, 561, 417, 399, 271 | |||||||
| 15′ | 76.776 | C55H92O24 | 1.46 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59766 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59009 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 16′ | 77.177 | C50H86O22 | 0.42 | 853 | 2-methylbutyric acid | +C10H16O3 | MS [M + FA − H]− 1083.55969 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1037.55469 | ||||||
| MS2 1019, 993, 935, 853, 835, 707, 663, 579, 561, 417, 399, 271 | |||||||
| 17′ | 77.521 | C55H92O24 | 1.08 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59729 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59583 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 18′ | 78.166 | C50H86O22 | 1.12 | 853 | 2-methylbutyric acid | +C10H16O3 | MS [M + FA − H]− 1083.56042 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1037.55444 | ||||||
| MS2 1019, 993, 935, 853, 835, 707, 663, 579, 561, 417, 399, 271 | |||||||
| 19′ | 78.756 | C55H92O24 | 0.92 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59717 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.58752 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 20′ | 79.008 | C54H92O24 | 1.27 | 853 | Isobutyric acid | +C14H22O5 | MS [M + FA − H]− 1169.59766 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1123.59155 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1079, 1035, 1023, 935, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 21′ | 79.452 | C55H92O24 | 1.17 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59729 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.58594 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 22′ | 79.881 | C50H84O22 | 0.94 | 853 | Tiglic acid | +C10H14O3 | MS [M + FA − H]− 1081.54468 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1035.53992 | ||||||
| MS2 991, 953, 935, 853, 835, 717, 679, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 23′ | 79.903 | C55H92O24 | 1.03 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59729 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59265 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 24′ | 80.623 | C55H94O24 | 0.80 | 853 | 2-methylbutyric acid | +C15H24O5 | MS [M + FA − H]− 1183.61243 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1137.60852 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1093, 1049, 1037, 991, 853, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 25′ | 80.774 | C54H92O24 | 1.35 | 853 | Isobutyric acid | +C14H22O5 | MS [M + FA − H]− 1169.59766 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1123.59155 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1079, 1035, 1023, 935, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 26′ | 81.148 | C55H92O24 | 1.22 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59705 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59314 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 27′ | 81.305 | C54H90O23 | 1.19 | 853 | 2-methylbutyric acid | +C14H20O4 | MS [M + FA − H]− 1151.58691 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1105.58093 | ||||||
| 3-hydroxy-2-methylenebutyric acid | MS2 1061, 1005, 973, 961, 917, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 28′ | 81.463 | C55H92O24 | 0.94 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59741 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59253 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 29′ | 82.021 | C55H94O24 | 0.72 | 853 | 2-methylbutyric acid | +C15H24O5 | MS [M + FA − H]− 1183.61243 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1137.60681 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1093, 1049, 1037, 991, 853, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 30′ | 82.154 | C54H90O23 | 1.17 | 853 | 2-methylbutyric acid | +C14H20O4 | MS [M + FA − H]− 1151.58679 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1105.58411 | ||||||
| 3-hydroxy-2-methylenebutyric acid | MS2 1061, 1005, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 31′ | 83.169 | C54H92O23 | 0.92 | 853 | Isobutyric acid | +C14H22O4 | MS [M + FA − H]− 1153.60205 |
| 2-methylbutyric acid | [M − H]− 1107.59619 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1063, 989, 905, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 32′ | 83.256 | C55H94O24 | 0.87 | 853 | 2-methylbutyric acid | +C15H24O5 | MS [M + FA − H]− 1183.61267 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1137.60510 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1093, 1049, 1037, 991, 853, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 33′ | 83.331 | C49H84O21 | 1.90 | 853 | 2-methylbutyric acid | +C9H12O2 | MS [M + FA − H]− 1053.55078 |
| Isobutyric acid | [M − H]− 1007.54419 | ||||||
| MS2 923, 905, 919, 835, 773, 671, 663, 579, 561, 417, 399, 271 | |||||||
| 34′ | 83.788 | C55H94O24 | 0.66 | 853 | 2-methylbutyric acid | +C15H24O5 | MS [M + FA − H]− 1183.61230 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1137.60889 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1093, 1049, 1037, 991, 853, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 35′ | 84.032 | C50H84O21 | 1.74 | 853 | 2-methylbutyric acid | +C10H14O2 | MS [M + FA − H]− 1065.55054 |
| Tiglic acid | [M − H]− 1019.54510 | ||||||
| MS2 937, 919, 835, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 36′ | 84.590 | C54H90O23 | 1.20 | 853 | Tiglic acid | +C14H20O4 | MS [M + FA − H]− 1151.58679 |
| isobutyric acid | [M − H]− 1105.58105 | ||||||
| 3-hydroxy-2-methylenebutyric acid | MS2 1061, 1005, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 37′ | 84.800 | C55H92O23 | 1.01 | 853 | Tiglic acid | +C15H22O4 | MS [M + FA − H]− 1165.60217 |
| 2-methylbutyric acid | [M − H]− 1119.59570 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1075, 1019, 1001, 937, 917, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 38′ | 84.863 | C49H84O21 | 1.66 | 853 | 2-methylbutyric acid | +C9H12O2 | MS [M + FA − H]− 1053.55042 |
| Isobutyric acid | [M − H]− 1007.54437 | ||||||
| MS2 923, 905, 919, 835, 773, 671, 663, 579, 561, 417, 399, 271 | |||||||
| 39′ | 84.632 | C53H90O23 | 0.63 | 853 | Isobutyric acid | +C13H20O4 | MS [M + FA − H]− 1139.58594 |
| Isobutyric acid | [M − H]− 1093.57764 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1049, 1005, 905, 835, 749, 661, 579, 561, 417, 399, 271 | ||||||
| 40′ | 85.717 | C50H84O21 | 0.48 | 853 | 2-methylbutyric acid | +C10H14O2 | MS [M + FA − H]− 1065.54919 |
| Tiglic acid | [M − H]− 1019.54413 | ||||||
| MS2 937, 919, 835, 661, 643, 579, 561, 417, 399, 271 | |||||||
| 41′ | 85.850 | C55H92O24 | 0.40 | 853 | Tiglic acid | +C15H22O5 | MS [M + FA − H]− 1181.59656 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1135.59082 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1091, 1047, 1035, 991, 935, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 42′ | 86.207 | C54H90O23 | 1.20 | 853 | Tiglic acid | +C14H20O4 | MS [M + FA − H]− 1151.58618 |
| isobutyric acid | [M − H]− 1105.58044 | ||||||
| 3-hydroxy-2-methylenebutyric acid | MS2 1061, 1005, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 43′ | 86.989 | C50H86O21 | 1.61 | 853 | 2-methylbutyric acid | +C10H16O2 | MS [M + FA − H]− 1067.56604 |
| 2-methylbutyric acid | [M − H]− 1021.55975 | ||||||
| MS2 937, 919, 853, 835, 663, 579, 561, 417, 399, 271 | |||||||
| 44′ | 87.502 | C55H90O23 | 0.41 | 853 | Tiglic acid | +C15H20O4 | MS [M + FA − H]− 1163.58655 |
| Tiglic acid | [M − H]− 1117.58057 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1099, 1073, 1017, 999, 937, 935, 917, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 45′ | 87.684 | C53H90O23 | 0.55 | 853 | Isobutyric acid | +C13H20O4 | MS [M + FA − H]− 1139.5863 |
| Isobutyric acid | [M − H]− 1093.57959 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1049, 1005, 905, 835, 749, 661, 579, 561, 417, 399, 271 | ||||||
| 46′ | 87.994 | C54H90O23 | 0.56 | 853 | Tiglic acid | +C14H20O4 | MS [M + FA − H]− 1151.58618 |
| isobutyric acid | [M − H]− 1105.58044 | ||||||
| 3-hydroxy-2-methylenebutyric acid | MS2 1061, 1005, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 47′ | 88.589 | C50H86O21 | 1.06 | 853 | 2-methylbutyric acid | +C10H16O2 | MS [M + FA − H]− 1067.56567 |
| 2-methylbutyric acid | [M − H]− 1021.55957 | ||||||
| MS2 937, 919, 853, 835, 663, 579, 561, 417, 399, 271 | |||||||
| 48′ | 88.674 | C54H92O23 | 0.58 | 853 | Isobutyric acid | +C14H22O4 | MS [M + FA − H]− 1153.60168 |
| 2-methylbutyric acid | [M − H]− 1107.59656 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1063, 989, 905, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 49′ | 89.303 | C55H92O23 | 0.29 | 853 | Tiglic acid | +C15H22O4 | MS [M + FA − H]− 1165.60132 |
| 2-methylbutyric acid | [M − H]− 1119.59595 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1075, 1037, 1001, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 50′ | 89.328 | C55H90O23 | 1.44 | 853 | Tiglic acid | +C15H20O4 | MS [M + FA − H]− 1163.58691 |
| Tiglic acid | [M − H]− 1117.58142 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1099, 1073, 1017, 937, 935, 891, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 51′ | 90.649 | C54H92O23 | 0.81 | 853 | Isobutyric acid | +C14H22O4 | MS [M + FA − H]− 1153.60193 |
| 2-methylbutyric acid | [M − H]− 1107.59668 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1063, 1007, 1005, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 52′ | 91.147 | C55H92O23 | 0.91 | 853 | Tiglic acid | +C15H22O4 | MS [M + FA − H]− 1165.60229 |
| 2-methylbutyric acid | [M − H]− 1119.59680 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1075, 1037, 1001, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 53′ | 91.797 | C55H94O23 | 0.59 | 853 | 2-methylbutyric acid | +C15H24O4 | MS [M + FA − H]− 1167.61743 |
| 2-methylbutyric acid | [M − H]− 1121.61243 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1077, 1021, 1019, 919, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 54′ | 91.855 | C54H90O23 | 1.39 | 853 | 2-methylbutyric acid | +C14H20O4 | MS [M + FA − H]− 1151.58716 |
| 3-hydroxy-2-methylbutyric acid | [M − H]− 1105.58069 | ||||||
| 3-hydroxy-2-methylenebutyric acid | MS2 1061, 1005, 961, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 55′ | 93.802 | C55H94O23 | 0.66 | 853 | 2-methylbutyric acid | +C15H24O4 | MS [M + FA − H]− 1167.61743 |
| 2-methylbutyric acid | [M − H]− 1121.61145 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1077, 1021, 1019, 919, 853, 835, 661, 635, 579, 561, 417, 399, 271 | ||||||
| 56′ | 94.849 | C55H92O23 | 1.09 | 853 | Tiglic acid | +C15H22O4 | MS [M + FA − H]− 1165.60242 |
| 2-methylbutyric acid | [M − H]− 1119.59680 | ||||||
| 3-hydroxy-2-methylbutyric acid | MS2 1075, 1037, 1001, 853, 835, 661, 635, 579, 561, 417, 399, 271 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Q.; Abdulla, R.; Kahar, G.; Aisa, H.A.; Li, C.; Xin, X. Mass Defect Filtering-Oriented Identification of Resin Glycosides from Root of Convolvulus scammonia Based on Quadrupole-Orbitrap Mass Spectrometer. Molecules 2022, 27, 3638. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27113638
Yin Q, Abdulla R, Kahar G, Aisa HA, Li C, Xin X. Mass Defect Filtering-Oriented Identification of Resin Glycosides from Root of Convolvulus scammonia Based on Quadrupole-Orbitrap Mass Spectrometer. Molecules. 2022; 27(11):3638. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27113638
Chicago/Turabian StyleYin, Qiang, Rahima Abdulla, Gulmira Kahar, Haji Akber Aisa, Chunting Li, and Xuelei Xin. 2022. "Mass Defect Filtering-Oriented Identification of Resin Glycosides from Root of Convolvulus scammonia Based on Quadrupole-Orbitrap Mass Spectrometer" Molecules 27, no. 11: 3638. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27113638