Bound Electron Enhanced Radiosensitisation of Nimorazole upon Charge Transfer
Abstract
:1. Introduction
2. Results
3. Discussion
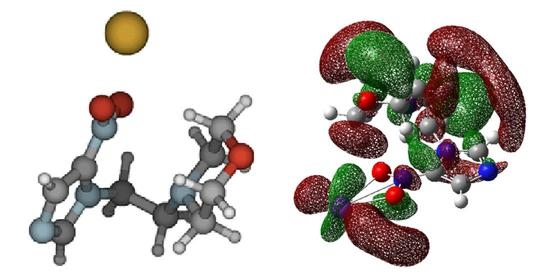
3.1. Parent Anion
3.2. NO2− Formation
3.3. Other Fragment Anions
4. Materials and Methods
4.1. The Crossed Molecular Beam Setup in the Lisbon Laboratory
4.2. Theoretical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Baumann, M.; Krause, M.; Overgaard, J.; Debus, J.; Bentzen, S.M.; Daartz, J.; Richter, C.; Zips, D.; Bortfeld, T. Radiation oncology in the era of precision medicine. Nat. Rev. Cancer 2016, 16, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Meißner, R.; Kočišek, J.; Feketeová, L.; Fedor, J.; Fárník, M.; Limão-Vieira, P.; Illenberger, E.; Denifl, S. Low-energy electrons transform the nimorazole molecule into a radiosensitiser. Nat. Commun. 2019, 10, 2388. [Google Scholar] [CrossRef] [PubMed]
- Meißner, R.; Feketeová, L.; Bayer, A.; Limão-Vieira, P.; Denifl, S. Formation of negative and positive ions in the radiosensitizer nimorazole upon low-energy electron collisions. J. Chem. Phys. 2021, 154, 074306. [Google Scholar] [CrossRef] [PubMed]
- Wardman, P. Some reactions and properties of nitro radical-anions important in biology and medicine. Environ. Health Perspect. 1985, 64, 309–320. [Google Scholar] [CrossRef]
- Oronsky, B.T.; Knox, S.J.; Scicinski, J. Six degrees of separation: The oxygen effect in the development of radiosensitizers. Transl. Oncol. 2011, 4, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Feketeová, L.; Albright, A.L.; Sørensen, B.S.; Horsman, M.R.; White, J.; O’Hair, R.A.J.; Bassler, N. Formation of radical anions of radiosensitizers and related model compounds via electrospray ionization. Int. J. Mass Spectrom. 2014, 365–366, 56–63. [Google Scholar] [CrossRef]
- Adams, G.E.; Flockhart, I.R.; Smithen, C.E.; Stratford, I.J.; Wardman, P.; Watts, M.E. Electron-Affinic Sensitization: VII. A Correlation between Structures, One-Electron Reduction Potentials, and Efficiencies of Nitroimidazoles as Hypoxic Cell Radiosensitizers. Radiat. Res. 2012, 178, AV183–AV189. [Google Scholar] [CrossRef]
- Ribar, A.; Fink, K.; Probst, M.; Huber, S.E.; Feketeová, L.; Denifl, S. Isomer Selectivity in Low-Energy Electron Attachment to Nitroimidazoles. Chem. Eur. J. 2017, 23, 12892–12899. [Google Scholar] [CrossRef]
- Meißner, R.; Feketeová, L.; Ribar, A.; Fink, K.; Limão-Vieira, P.; Denifl, S. Electron Ionization of Imidazole and Its Derivative 2-Nitroimidazole. J. Am. Soc. Mass Spectrom. 2019, 30, 2678–2691. [Google Scholar] [CrossRef]
- Itälä, E.; Tanzer, K.; Granroth, S.; Kooser, K.; Denifl, S.; Kukk, E. Fragmentation patterns of 4(5)-nitroimidazole and 1-methyl-5-nitroimidazole—The effect of the methylation. J. Mass Spectrom. 2017, 52, 770–776. [Google Scholar] [CrossRef]
- Itälä, E.; Myllynen, H.; Niskanen, J.; González-Vázquez, J.; Wang, Y.; Ha, D.T.; Denifl, S.; Kukk, E. Controlling NO Production Upon Valence Ionization of Nitroimidazoles. J. Phys. Chem. A 2019, 123, 3074–3079. [Google Scholar] [CrossRef] [PubMed]
- Tanzer, K.; Feketeová, L.; Puschnigg, B.; Scheier, P.; Illenberger, E.; Denifl, S. Reactions in Nitroimidazole Triggered by Low-Energy (0–2 eV) Electrons: Methylation at N1-H Completely Blocks Reactivity. Angew. Chem. Int. Ed. 2014, 53, 12240–12243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandeti, S.; Feketeová, L.; Reddy, T.J.; Abdoul-Carime, H.; Farizon, B.; Farizon, M.; Märk, T.D. Binding preference of nitroimidazolic radiosensitizers to nucleobases and nucleosides probed by electrospray ionization mass spectrometry and density functional theory. J. Chem. Phys. 2019, 150, 014302. [Google Scholar] [CrossRef]
- Khreis, J.M.; Pandeti, S.; Feketeová, L.; Denifl, S. High-energy collision-induced dissociation of radiosensitizer anions: Nimorazole and metronidazole. Int. J. Mass Spectrom. 2018, 431, 1–7. [Google Scholar] [CrossRef]
- Pandeti, S.; Feketeová, L.; Reddy, T.J.; Abdoul-Carime, H.; Farizon, B.; Farizon, M.; Märk, T.D. Nitroimidazolic radiosensitizers investigated by electrospray ionization time-of-flight mass spectrometry and density functional theory. RSC Adv. 2017, 7, 45211–45221. [Google Scholar] [CrossRef] [Green Version]
- Ferreira da Silva, F.; Almeida, D.; Antunes, R.; Martins, G.; Nunes, Y.; Eden, S.; Garcia, G.; Limão-Vieira, P. Electron transfer processes in potassium collisions with 5-fluorouracil and 5-chlorouracil. Phys. Chem. Chem. Phys. 2011, 13, 21621–21629. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.; Probst, M.; Maihom, T.; García, G.; Limão-Vieira, P. Selective Bond Excision in Nitroimidazoles by Electron Transfer Experiments. J. Phys. Chem. A 2019, 123, 4068–4073. [Google Scholar] [CrossRef]
- Mendes, M.; García, G.; Bacchus-Montabonel, M.C.; Limão-Vieira, P. Electron transfer induced decomposition in potassium–nitroimidazoles collisions: An experimental and theoretical work. Int. J. Mol. Sci. 2019, 20, 6170. [Google Scholar] [CrossRef] [Green Version]
- Sonveaux, P.; Jordan, B.F.; Gallez, B.; Feron, O. Nitric oxide delivery to cancer: Why and how? Eur. J. Cancer 2009, 45, 1352–1369. [Google Scholar] [CrossRef]
- Almeida, D.; Ferreira da Silva, F.; García, G.; Limão-Vieira, P. Selective bond cleavage in potassium collisions with pyrimidine bases of DNA. Phys. Rev. Lett. 2013, 110, 023201. [Google Scholar] [CrossRef]
- Cunha, T.; Mendes, M.; Ferreira da Silva, F.; Eden, S.; García, G.; Limão-Vieira, P. Communication: Site-selective bond excision of adenine upon electron transfer. J. Chem. Phys. 2018, 148, 021101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NIST Chemistry Webbook. Available online: http://webbook.nist.gov/chemistry (accessed on 6 May 2022).
- Kleyn, A.W.; Moutinho, A.M.C. Negative ion formation in alkali-atom–molecule. J. Phys. B At. Mol. Opt. Phys. 2001, 4075, R1–R44. [Google Scholar] [CrossRef]
- Almeida, D.; Ferreira da Silva, F.; Eden, S.; García, G.; Limão-Vieira, P. New fragmentation pathways in K-THF collisions as studied by electron-transfer experiments: Negative ion formation. J. Phys. Chem. A 2014, 118, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Ferreira da Silva, F.; García, G.; Limão-Vieira, P. Dynamic of negative ions in potassium-D-ribose collisions. J. Chem. Phys. 2013, 139, 114304. [Google Scholar] [CrossRef] [Green Version]
- Almeida, D.; Ferreira Da Silva, F.; Kopyra, J.; García, G.; Limão-Vieira, P. Anion formation in gas-phase potassium-uridine collisions. Int. J. Mass Spectrom. 2014, 365–366, 243–247. [Google Scholar] [CrossRef]
- Lozano, A.I.; Kumar, S.; Kerkeni, B.; García, G.; Limão-Vieira, P. Methanol Negative Ion Fragmentation Probed in Electron Transfer Experiments. J. Phys. Chem. A 2022, 126, 1076–1084. [Google Scholar] [CrossRef]
- Almeida, D.; Kinzel, D.; Ferreira da Silva, F.; Puschnigg, B.; Gschliesser, D.; Scheier, P.; Denifl, S.; García, G.; González, L.; Limão-Vieira, P. N-site de-methylation in pyrimidine bases as studied by low energy electrons and ab initio calculations. Phys. Chem. Chem. Phys. 2013, 15, 11431. [Google Scholar] [CrossRef] [Green Version]
- Ferreira da Silva, F.; Meneses, G.; Ingólfsson, O.; Limão-Vieira, P. Side chain effects in reactions of the potassium-tyrosine charge transfer complex. Chem. Phys. Lett. 2016, 662, 19–24. [Google Scholar] [CrossRef]
- Mendes, M.; Pamplona, B.; Kumar, S.; da Silva, F.F.; Aguilar, A.; García, G.; Bacchus-Montabonel, M.C.; Limão-Vieira, P. Ion-pair formation in neutral potassium-neutral pyrimidine collisions: Electron transfer experiments. Front. Chem. 2019, 7, 264. [Google Scholar] [CrossRef]
- da Silva, F.F.; Cunha, T.; Rebelo, A.; Gil, A.; Calhorda, M.J.; García, G.; Ingólfsson, O.; Limão-Vieira, P. Electron-Transfer-Induced Side-Chain Cleavage in Tryptophan Facilitated through Potassium-Induced Transition-State Stabilization in the Gas Phase. J. Phys. Chem. A 2021, 125, 2324–2333. [Google Scholar] [CrossRef]
- da Silva, F.F.; Lança, M.; Almeida, D.; García, G.; Limão-Vieira, P. Anionic fragmentation of glycine upon potassium-molecule collisions. Eur. Phys. J. D 2012, 66, 78. [Google Scholar] [CrossRef]
- Silva, F.F.; Mendes, M.; García, G.; Limão-Vieira, P. Radiation in Bioanalysis, Spectroscopic Techniques and Theoretical Methods; Pereira, A.S., Tavares, P., Limão-Vieira, P., Eds.; Springer: Cham, Switzerland, 2019; pp. 329–348. [Google Scholar]
- Kleyn, A.; Los, J.; Gislason, E.A. Vibronic Coupling At Intersections of Covalent and Ionic States. Phys. Rep. 1982, 90, 1–71. [Google Scholar] [CrossRef]
- Kumar, S.; Kilich, T.; Łabuda, M.; García, G.; Limão-Vieira, P. Anionic states of C6Cl6 probed in electron transfer experiments. Phys. Chem. Chem. Phys. 2022, 24, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Antunes, R.; Martins, G.; Eden, S.; Ferreira da Silva, F.; Nunes, Y.; Garcia, G.; Limão-Vieira, P. Electron transfer-induced fragmentation of thymine and uracil in atom–molecule collisions. Phys. Chem. Chem. Phys. 2011, 13, 15657. [Google Scholar] [CrossRef] [PubMed]
- Antunes, R.; Almeida, D.; Martins, G.; Mason, N.J.; Garcia, G.; Maneira, M.J.P.; Nunes, Y.; Limão-Vieira, P. Negative ion formation in potassium–nitromethane collisions. Phys. Chem. Chem. Phys. 2010, 12, 12513–12519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meneses, G.; Widmann, C.; Cunha, T.; Gil, A.; Ferreira Da Silva, F.; Calhorda, M.J.; Limão-Vieira, P. Unravelling the dissociation pathways of acetic acid upon electron transfer in potassium collisions: Experimental and theoretical studies. Phys. Chem. Chem. Phys. 2017, 19, 1083–1088. [Google Scholar] [CrossRef]
- Su, X.; Cheng, X.; Meng, C.; Yuan, X. Quantum chemical study on nitroimidazole, polynitroimidazole and their methyl derivatives. J. Hazard. Mater. 2009, 161, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Cunha, T.; Mendes, M.; Ferreira da Silva, F.; Eden, S.; García, G.; Bacchus-Montabonel, M.C.; Limão-Vieira, P. Electron transfer driven decomposition of adenine and selected analogs as probed by experimental and theoretical methods. J. Chem. Phys. 2018, 148, 134301. [Google Scholar] [CrossRef] [Green Version]
- Ferreira da Silva, F.; Matias, C.; Almeida, D.; García, G.; Ingólfsson, O.; Flosadóttir, H.D.; Ómarsson, B.; Ptasinska, S.; Puschnigg, B.; Scheier, P.; et al. NCO-, a key fragment upon dissociative electron attachment and electron transfer to pyrimidine bases: Site selectivity for a slow decay process. J. Am. Soc. Mass Spectrom. 2013, 24, 1787–1797. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Ravanat, J.L. Oxidatively generated base damage to cellular DNA. Free Radic. Biol. Med. 2010, 49, 9–21. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Ravanat, J.L. Measurement of oxidatively generated base damage in cellular DNA. Mutat. Res. 2011, 711, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Dumont, E.; Monari, A. Understanding DNA under oxidative stress and sensitization: The role of molecular modeling. Front. Chem. 2015, 3, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravanat, J.-L.; Cadet, J.; Douki, T. Oxidatively Generated DNA Lesions as Potential Biomarkers of In Vivo Oxidative Stress. Curr. Mol. Med. 2012, 12, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Regeta, K.; Kumar, S.; Cunha, T.; Mendes, M.; Lozano, A.I.; Pereira, P.J.S.; García, G.; Moutinho, A.M.C.; Bacchus-Montabonel, M.C.; Limão-Vieira, P. Combined Experimental and Theoretical Studies on Electron Transfer in Potassium Collisions with CCl4. J. Phys. Chem. A 2020, 124, 3220–3227. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Blaudeau, J.P.; McGrath, M.P.; Curtiss, L.A.; Radom, L. Extension of Gaussian-2 (G2) theory to molecules containing third-row atoms K and Ca. J. Chem. Phys. 1997, 107, 5016–5021. [Google Scholar] [CrossRef]
- McLean, A.D.; Chandler, G.S. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z = 11–18. J. Chem. Phys. 1980, 72, 5639–5648. [Google Scholar] [CrossRef]
- Petersilka, M.; Gossmann, U.J.; Gross, E.K.U. Excitation Energies from Time-Dependent Density-Functional Theory. Phys. Rev. Lett. 1996, 76, 1212–1215. [Google Scholar] [CrossRef] [Green Version]
- Wardman, P. Chemical Radiosensitizers for Use in Radiotherapy. Clin. Oncol. 2007, 19, 397–417. [Google Scholar] [CrossRef]
- Wardman, P. The mechanism of radiosensitization by electron-affinic compounds. Radiat. Phys. Chem. 1987, 30, 423–432. [Google Scholar] [CrossRef]







| Mass (u) | Mass (u) [2,3] | Assignment | Major Source of Fragmentation | |
|---|---|---|---|---|
| 26, 27 | 26 | CN−, 13CN− | 4-nitromidazole ring | |
| 30 | − | NO− | 4-nitromidazole ring | |
| 38 | − | C2N−/C3H2− | ||
| 39 | − | C2HN−/C3H3− | ||
| 40 | − | C2H2N− | ||
| 41 | − | C2H3N−/CHN2− | ||
| 42 | 42 | CNO− | 4-nitromidazole ring | |
| 43 | − | CHNO− | ||
| 46, 47, 48 | 46, 48 | NO2−, 15NO2−, 14N18OO− | 4-nitromidazole ring | |
| 52 | − | C3H2N− | ||
| 56 | − | C2H2NO−/C3H6N− |  | 4-nitromidazole ring |
| 64 | − | C3N2− | ||
| 65 | 65 | C3HN2− | ||
| 66 | − | C3H2N2− | ||
| 67 | 67 | C3H3N2− | ||
| 68 | − | C3H4N2− | ||
| 69 | − | C3H5N2− | ||
| 79 | − | C4H3N2− | ||
| 80 | − | C4H2NO− | ||
| 81 | − | C3HN2O−/C4H3NO− |  | morpholine ring |
| 82 | 82 | C3H2N2O−/C4H4NO− | ||
| 83 | 83 | C3H3N2O−/C4H5NO− | ||
| 84 | − | C3H4N2O−/C4H6NO− | ||
| 86 | − | C4H8NO− | ||
| 93 | − | C4HN2O− | ||
| 94 | − | C3N3O− | ||
| 95 | − | C4H3N2O− | ||
| 96 | − | C3H2N3O− | ||
| 97 | 97 | C3H3N3O− | ||
| 108 | − | C4H2N3O− | ||
| 109 | − | C4HN2O2− | ||
| 111 | − | C4H3N2O2− | ||
| 112 | 112 | C3H2N3O2− | ||
| 113 | − | C4H5N2O2− | ||
| 114 | − | C3H4N3O2− | ||
| 126 | − | C4H4N3O2− | ||
| 134 | − | C5N3O2− | ||
| 138 | − | C5H4N3O2− | ||
| 139 | − | C4H3N4O2− | ||
| − | 178 | (NIMO–NO2–2H)− | ||
| 196 | 196 | (NIMO–NO)− | ||
| − | 208 | (NIMO–H2O)− | ||
| − | 209 | (NIMO–OH)− | ||
| 210 | 210 | (NIMO–O)− | ||
| 225 | 225 | (NIMO–H)− | ||
| 226, 227, 228, 229 | 226 | NIMO•– and its isotopes | ||
| K+ Energy Loss Feature (eV) | VEA (eV) | Calculated VE of MO (eV) | Assignment | EA Resonances [2,3] |
|---|---|---|---|---|
| 4.74 ± 0.07 | −0.40 ± 0.07 | 4.81 | LUMO + 50 | 0.41 |
| 6.11 ± 0.04 | −1.77 ± 0.04 | 5.69 | LUMO + 60 | 1.49, 1.93 |
| 7.21 ± 0.03 | −2.87 ± 0.03 | 7.01 | LUMO + 70 | 2.97 |
| 8.17 ± 0.06 | −3.83 ± 0.06 | 8.28 | LUMO + 80 | 3.53, 3.6 |
| 9.55 ± 0.11 | −5.21 ± 0.11 | 9.17 | LUMO + 90 | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Ben Chouikha, I.; Kerkeni, B.; García, G.; Limão-Vieira, P. Bound Electron Enhanced Radiosensitisation of Nimorazole upon Charge Transfer. Molecules 2022, 27, 4134. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27134134
Kumar S, Ben Chouikha I, Kerkeni B, García G, Limão-Vieira P. Bound Electron Enhanced Radiosensitisation of Nimorazole upon Charge Transfer. Molecules. 2022; 27(13):4134. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27134134
Chicago/Turabian StyleKumar, Sarvesh, Islem Ben Chouikha, Boutheïna Kerkeni, Gustavo García, and Paulo Limão-Vieira. 2022. "Bound Electron Enhanced Radiosensitisation of Nimorazole upon Charge Transfer" Molecules 27, no. 13: 4134. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27134134