1. Introduction
Lung cancers are the second most occurring cancers and the primary cause of cancer-related death among both men and women worldwide [
1]. In 2018, out of 9.6 million cancer-related deaths, 1.76 million were caused by lung cancers [
2]. Non-Small Cell Lung Cancer (NSCLC) accounts for 85% of all lung cancers [
3], and adenocarcinoma is the principal subtype makeup for 75–85% of lung cancer and related deaths [
4].
Rapid metastasis of lung adenocarcinomata [
5] are primarily responsible for their late diagnosis, significantly affecting patient survival [
6]. Common causes of lung cancers include smoking, air pollution, occupational exposure, and genetics [
6]. However, diet also plays a crucial chemoprophylactic role in lung cancers [
7,
8,
9]. It is more evident from the increasing number of novel anticancer compounds discovered in common foods and drinks [
10]. Besides rapid metastasis, the aggressive chemotherapeutic regimen often associated with late-stage lung cancer treatment also contributes to poor patient survival [
11]. Natural products used as adjuvant therapy have promoted patient survival and quality of life [
12,
13]. These have created a demand for natural chemopreventive, chemoprotective, and adjuvant compounds in cancer treatment [
3,
12,
13,
14].
Natural products are an abundant source of novel therapeutics [
14]. More than 67% of novel anticancer and antitumor drugs are natural compounds or their analog [
14,
15], and more than 200 compounds and derivatives are at different stages of drug development and trials [
15]. Chicory (
Cichorium intybus L.) is a widely distributed plant used as medicine, food, drink, and fodder [
16]. It has been the subject of many pharmacological studies [
16]. Recently, we reviewed the cytotoxic of Chicory [
16], where 31 out of 87
C. intybus metabolites possess anticancer, antitumor, and related bioactivities. Lactucin is a phytometabolite from Chicory, phytochemically categorized as bitter Sesquiterpene Lactones (SLs). SLs have been a subject of interest in cancer research for decades, and many have reached clinical trials [
15]. As indicated in Ghantous, Gali-Muhtasib, Vuorela, Saliba, and Darwiche [
15] reviews, SLs bioactivity is strictly linked to three conserved structural features, e.g., (i) alkylating center reactivity, (ii) side chain and lipophilicity, and (iii) molecular geometry and electronic features. Since many of the structural features that lactucin shares with other SLs have shown antitumor bioactivity against NSCLC [
15], Lactucin will likely possess these properties.
Some plants of the Asteraceae family commonly synthesize Lactucin. It is one of the ingredients of lactucarium, a milky white liquid secreted by several lettuce species, e.g.,
Lactuca serriola,
L. saligna,
L. viminea,
L. glareosa,
L. sativa, [
17,
18,
19], etc. In 1970 Chicory root water extract was reported as a light-sensitive, highly potent antimalarial compound. Later Bischoff et al. [
20] attributed it to Lactucin, making Chicory the most famous source of Lactucin. Besides antimalarial properties, Lactucin also exerts or potentially exerts anti-inflammatory [
21], sedatives [
17], anti-adipogenic [
22], and anthelmintics [
23] effects. Zhang et al. [
24] reported Lactucin induces apoptosis and sub-G
1 cell cycle arrest in HL-60 (human leukemia cancer) cells. Ren et al. [
25] reported similar effect on KB (human epidermoid carcinoma; IC
50 = 75 μM), and Bel 7402 (human hepatocellular carcinoma; IC
50 = 55 μM) cells. Apart from these reports, the anticancer effect of Lactucin, especially on lung cancer, remains largely unexplored. A structural activity study (SAR) by Ren, et al. [
25] revealed the importance of an ester group (γ-butyrolactone) and one exocyclic methylene group for the antitumor activity of Lactucin-like Guanolides. Wang, et al. [
22] reported that Lactucin inhibits adipogenesis by downregulating the JAK2/STAT3 signaling pathway and subsequent clonal expansion. However, the cellular target protein of Lactucin and the affected pathway in antitumor activity is still unknown. Herein, we evaluated the anticancer potency of Lactucin using A549 and H2347 lung adenocarcinoma cell lines in vitro to identify interacting proteins and underlying molecular mechanisms.
3. Discussion
In the present experiment, we observed that Lactucin has significant antiproliferative, G
0/G
1 cell cycle arrest, and apoptosis-inducing properties on A549 and H2347 lung adenocarcinoma, not on MRC-5 normal lung cells. Firstly, Lactucin-induced time and dose-dependent inhibition of A549 and H2347 cells were observed, and then their IC
50 values were determined to be 79.87 μM and 68.85 μM, respectively. According to WHO [
28], for L-6 (rat skeletal myoblast) cells, compounds with an IC
50 above 90 μM are not cytotoxic, IC
50 between 2–80 μM is moderately cytotoxic, and IC
50 below 2 μM is cytotoxic. For natural compounds, Ren, et al. [
25] described an IC
50 below 100 μM to be cytotoxic and reported Lactucin inhibits KB and Bel 7402 cells with an IC
50 value of 75 μM and 55 μM, respectively. Lima, et al. [
29] considered IC
50 of <40 µg/mL and <4 µg/mL for plant extract and pure compounds respectively to be cytotoxic. According to American National Cancer Institute (NCI), except for fibroblast, an IC
50 of ≤30 µg/mL of plant extract is cytotoxic, but they did not specify pure natural compounds. IC
50 values we observed in adenocarcinomas can thus be considered cytotoxic. The LC3-II and LC3-II/LC3-I ratio increase is a classic sign of autophagy [
30]. However, this is not always the case due to the lower sensitivity of LC3-I than LC3-II in WB and the higher degradation rate of LC3-II in the presence of lysosomal protease inhibitor [
30,
31]. We observed a dose-dependent increase of LC3-II in H2347 and LC3-I in A549 and H2347 cells. It was probably due to the concomitant increase in LC3 production and LC3I to LC3II conversion, rapid degradation of LC3-II, or lower detection of LC3-I in WB. In conclusion, LC3 WB results didn’t sufficiently indicate that Lactucin directly induces autophagy. LC3-I and LC3-II increase was probably due to autophagosome increase by some other means [
30].
We found Lactucin induces G
0/G
1 cell cycle arrests from cell cycle analysis. Specific cell cycle-related proteins, such as cyclins and CDKs, positively upregulate the cell cycle, whereas CDKs inhibitors stop the Cyclins and CDKs unit assembly [
32]. In this experiment, we observed the downregulation of Cyclin B1, Cyclin D1, CDK2, and CDK4, and the upregulation of p21 and p53. Downregulation of Cyclin D1 and upregulation of p53 were associated with G
0/G
1 cell cycle arrest and apoptosis [
32], which coincides with our findings. Lactucin-induced inhibition of cell proliferation was also reported by Zhang, et al. [
24] in human HL-60 leukemia cancer cells and by Wang, et al. [
22] in mouse 3T3-L1 fibroblast cells. Though in 3T3-L1 G
0/G
1 cell cycle arrest was reported, [
22] in HL-60 cells, Sub-G
1 cell cycle arrest was reported [
24].
We also observed an increase in early and late apoptosis in A549 and H2347 cells induced by Lactucin. Lactucin induces apoptosis by upregulating the expression of mitochondrial apoptosis-related proteins, such as c-Caspase, c-PARP, and Bax, while downregulating the expression of Bcl-2. Zhang, et al. [
24] reported Lactucin induces apoptosis in HL-60 cells by swelling up mitochondria and endoplasmic reticulum (ER) on the transition electron microscope (TEM). Jang, et al. [
33] reported that Lactucin induces ROS-mediated apoptosis in human renal cancer cell Caki-1 by downregulating Bcl-2 expression and CFLARL stability where Bcl-2 downregulation was at a transcriptional level caused by inactivation of the NF-κB pathway. However, we observed concurrent upregulation of PTEN and downregulation of Akt, MEK, and ERK phosphorylation narrowed down the Lactucin-influenced NSCLC pathway to PI3K/Akt and MAPK/ERK. McCubrey, et al. [
34] suggested that ERK can activate the NF-κB transcription factor (nuclear factor immunoglobulin κ chain enhancer-B cell) by phosphorylating and activating inhibitor κB kinase (IKK) through an indirect mechanism. We also performed an ABPP assay on A549 protein lysate using a Lactucin-Propargylamine probe. Structural activity relationship (SAR) studies by Ren, et al. [
25] revealed that in SL, the position 8 ester group (γ-butyrolactone) and the methylene group at exocyclic position 11 (α), play a significant role in antitumor activities of Lactucin-like guaianolides (
Figure 5A) [
16]. α methylene γ lactone, the’ enone’ or unsaturated carbonyl system (O=C–C=CH
2) was also reported to increase the toxicity towards tumor cells [
15]. While synthesizing the Lactucin-Propargylamine probe, we avoided the 8-ester group and 11 methylene group positions mentioned earlier. However, adding the alkynyl group to 11 (α), exocyclic methylene didn’t hinder its cytotoxicity against A549 cells (
Figure 5B,C). It also suggests that Lactucin may simultaneously exert its activity through different groups. Proteins isolated using this probe were analyzed in LC-MS/MS and combined with WB results for functional enrichment. In the DAVID functional enrichment of western blot and ABPP peptides, four were identified as key enzymes in central carbon metabolism in the cancer cell (
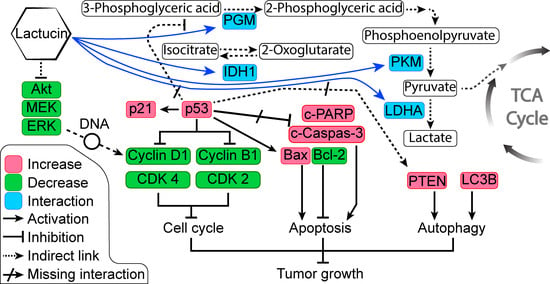
Figure 6C). Overexpressed and modified central carbon metabolism enzymes are essential to support the exponential growth of the metastasized tumor. Regulating their expression will limit the available energy and thus control or seize tumor growth.
In anticancer therapy, oncogenes and tumor suppressors are two likely targets for inhibiting cancer cells [
26]. From the KEGG pathway of NSCLEC by Kanehisa Laboratories, we know that in the case of NSCLC like A549 or H2347, we know the potential oncogene and tumor suppressors targets. DAVID enrichment results showed that in the NSCLC, Lactucin downregulates MEK-ERK pathway, resulting in the downregulation of CyclinD1, CDKs, and upregulation of the p53. p53, in turn, upregulates p21 and Bax. It doesn’t show interaction with any of the upstream components of NSCLC. When DAVID enrichment for the KEGG pathway “Central Carbon Metabolism in Cancer” was plotted, it showed four Lactucin interacting enzymes besides the western blot findings. Downregulation of MAPK/ERK observed in
Figure 6C also lowers the expression of c-Myc, a proto-oncogene involved in cell cycle progression, apoptosis, and cellular transformation. Low c-Myc inhibits uncontrolled DNA synthesis associated with cancer and thus lowers the expression of PGM, PKM, and LDHA, and eventually downregulates carbon consumption needed to sustain malignancy. Lactucin also binds to PDH, reducing its concentration, and decreasing Pyruvate to Acetyl CoA conversion, thus downregulating the TCA cycle and lactate production. Thus, Lactucin inhibits cell proliferation and simultaneously increases cell cycle arrest in lung adenocarcinoma by reducing the MAPK/ERK and lowering carbon metabolism-related enzyme availability.
4. Materials and Methods
Firstly, we tested the Lactucin for its cytotoxicity, cell cycle inhibiting, and apoptosis-inducing properties on lung adenocarcinoma cell lines. Then, its effect on cell cycle, apoptosis, and other related protein expression, was evaluated by WB analysis. Finally, we constructed a biotinylated Lactucin probe to extract Lactucin-interacting proteins, identified them using LC-MS/MS, and elucidated the molecular mechanisms.
4.1. Cell Materials
In the present study, we observed the effect of Lactucin (Shanghai Yuanye Biological Technology Co., Ltd., Shanghai, China) on two lung adenocarcinoma cell lines, namely, A549 (Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China), and H2347 (Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China), as well as one normal lung cell line MRC-5 (National Infrastructure of Cell Line Resource, Beijing, China).
4.2. Cell Culture
Frozen (liquid-vapor) cell lines were cultured in complete growth media [A549 and H2347 cells in Roswell Park Memorial Institute 1640 (RPMI-1640) with 10% Fetal Bovine Serum (FBS) (Biological Industries, Kibbutz Beit Haemek, Israel) and 1% Penicillin/Streptomycin (Caisson Laboratories, Smithfield, VA, USA); MRC-5 in Minimum Essential Medium (MEM HyClone, Chicago, IL, USA), with 10% FBS, 1% Penicillin/Streptomycin, and 1% Nonessential Amino acids (10 mM 100×, Solarbio Biotechnology, Beijing, China) (EMEM)] [
26]. For freezing A549, H2347, and MRC-5 cell line, 10% Dimethyl sulfoxide (DMSO, MP Biomedicals, Shanghai, China) in FBS, RPMI-1640, and MEM complete growth medium, respectively, was used. Adherent cells were detached using 1–3 mL of 0.25% Trypsin EDTA (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) for 2–4 min at room temperature (RT).
4.3. Cell Viability Assay
Subconfluent cells (<90%) were aseptically trypsinized and diluted to 5 × 104 cell/mL using complete growth media RPMI-1640 (for A549 and H2347) or EMEM (for MRC-5). Cells were then seeded aseptically in 96 welled plates at 1 × 104 cells/well (200 µL/well) and incubated for 12 h at 37 °C in a 5% CO2 incubator before dosing. After 12 h, we exposed the cells to different concentrations of Lactucin for 24 h. All Lactucin concentrations were prepared using complete growth media and contained equal vehicle volume DMSO (v/v). Cell viability was determined using a MTT cell proliferation kit (M1020, Solarbio Biotechnology, Beijing, China) following the manufacturer’s instructions. UV absorbance was scanned at 490 nm using a Spark microtitration plate reader (Tecan, Männedorf, Switzerland). Each experiment was repeated three times, the viability of the control group was set to 100%, and cell viability and IC50 values were calculated.
4.4. DNA Content/Cell Cycle Assay
Subconfluent A549 and H2347 cells (<90%) were cultured at 1 × 105 cell/mL concentration using complete growth media RPMI-1640 for 12 h at 37 °C in 5% CO2. Cells were then treated with respective IC50 Lactucin solution or equivalent volume DMSO (v/v) solution and incubated for 24 h. After treatment, cells were trypsinized, washed, and fixed for 24 h. Fixed cells were washed and stained using a Propidium Iodide (PI) Flow Cytometry kit (Abcam, Cambridge, UK) following manufacturer instructions. The DNA contents were scanned using a flow cytometer (CytoFLEX, Beckman Coulter Inc., Miami, FL, USA).
4.5. Apoptosis Assay
Subconfluent A549 and H2347 cells (<90%) were cultured at 1 × 105 cell/mL concentration using complete growth media RPMI-1640 for 12 h at 37 °C in 5% CO2. Cells were then treated with respective IC50 Lactucin solution or equivalent volume DMSO (v/v) solution and incubated for 24 h. After treatment, cells were trypsinized and washed. Following manufacturer instructions, cells were double-stained using the FITC Annexin V Apoptotic kit (BD Pharmingen, San Diego, CA, USA). Samples were filtered (70 μM Nylon cell strainer, Falcon, Corning, Durham, NC, USA) and scanned using a flow cytometer.
4.6. Protein Extraction
5 × 106 cell/mL untreated [A549 for Co-IP] and treated (Lactucin and DMSO in A549 and H2347 for WB) cells were washed (1× PBS chilled), scraped (cell scraper, Corning, Durham, NC, USA), collected in 1.5 mL centrifuge tubes, and then washed again twice (1× PBS chilled). Between each washing, cells were centrifuged at 600× g for 5 min at 4 °C. After the final centrifuge, 300–500 µL of chilled modified RIPA buffer (1% PMSF + 1% SDS in RIPA) was added to each tube, briefly vortexed and then homogenized using probe sonicator at 30% power of 3 × 5 s with a 10 s gap in between. Homogenized cells were then left to chill on ice for 1 h. After 1 h, cells were centrifuged at 14000× g at 4 °C for 10 min, and intact cells and nuclear materials were separated by pipetting the supernatant into new centrifuge tubes and stored at −20 °C. The protein concentrations were determined using the Bicinchoninic Acid (BCA) protein assay kit (Solarbio Biotechnology, Beijing, China) following manufacturer instructions. UV absorbance was scanned at 562 nm using a microtitration plate reader.
4.7. Western Blot Assay
Western blot was performed using the method adopted from Lu. et al. [
26]. A549 and H2347 cells protein (10 µg/lane) along with a pre-stained page ruler (5 µg/lane) were resolved using 8, 10, and 12% SDS-PAGE, along with pre-stained protein ladder (#26617; Thermo Fisher Scientific, Carlsbad, CA, USA) and transferred to polyvinylidene fluoride (PVDF) transfer membrane (Immobilon
®-P, Billerica, MA, USA) by wet transfer at 350 mA for 70–110 min (depending on targeted proteins molecular weight). PVDF membranes were washed thrice using Tris-buffered saline with 0.1% Tween
® 20 Detergent (TBST) (10 min each) and blocked using 5% Bovine Serum Albumin (BSA) in TBST (10 mM Tris-HCl, 150 mM NaCl, 0.1% tween 20) at room temperature (RT) for 1 h. The PVDF was then washed thrice using TBST (10 min each), cut, and probed using primary antibodies at 4 °C overnight. The primary antibodies include anti-Akt (AA326), anti-Bax (AB026), anti-cleaved Caspase-3 (AC033), anti-CDK-4 (AC251-1), anti-Cyclin D1 (AC853-1), anti-p53 (AF0255), anti-ERK1/2 (AF1051), anti-MEK1/2 (AF1057), anti-cleaved-PARP-1 (AF1567), anti-mTOR (AF1648), anti-Ki67 (AF1738), anti-p-ERK (AF1891), anti-p-Akt1/2/3(Thr 308) (AF5734), anti-p21 (AP021-1), anti-PTEN (AP686) from Beyotime Biotechnology (Shanghai, China); anti-LC3B (NB100-2220) from Novus Biologicals (Littleton, CO, USA); anti-β-actin (SC47778), anti-Cdk-2 (SC6248), anti-Cyclin B1 (SC245), anti- p-MEK1/2 (SC81503), anti-Bcl-2 (SC7882) from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Next, the PVDF was washed thrice using TBST (10 min each) and probed with appropriate goat anti-mouse (ab150113) or anti-rabbit (ab97051) secondary antibodies (Abcam, Cambridge, UK) for 1 h at RT. Finally, the PVDF was washed (thrice for 10 min each), exposed using luminol reagents (Millipore, Billerica, MA, USA), and photographed using the CLiNX Chemiluminescence imaging system (Shanghai, China).
4.8. Synthesis of Lactucin Probe
To synthesize the biotinylated Lactucin probe, Lactucin was conjugated with an alkynyl group through Michael’s addition reaction,
Figure 6A. To a solution of lactucin (5.0 mg, 0.018 mM) in dry pyridine (0.3 mL), Propargylamine (1.5 mg, 0.027 mM) (inno-chem, Beijing, China) was added and stirred at 0 °C for 36 h under N
2 air. The reaction was monitored by thin-layer chromatography (TLC). The solvent was concentrated and purified through column chromatography (Dichloromethane-CH
3OH, 15:1) and dried. For structural confirmation, the probe was analyzed using
1H-NMR at 600 MHz (Oxford NMR AS600, Abingdon, UK) in deuterated DMSO (d-DMSO, Merck KGaA, Darmstadt, Germany),
Figure 5A. The yield of the probe was estimated using HPLC.
4.9. Cytotoxicity Test and Protein Labelling by Lactucin Probe
Cytotoxicity of the Lactucin probe and Propargylamine was examined by incubating A549 cells with incriminating (0 to 150 μM) concentrations and incubation periods (12, 24, and 48 h) of each. The total A549 protein lysate was prepared using the method described in
Section 4.6. The Lactucin binding proteins were labeled and pulled out using a biotinylated Lactucin probe following the ABPP method adopted from the method described by Speers and Cravatt [
35]. Two 400 µL of protein lysate (2 mg/mL in PBS) were aliquoted into a 1.5mL microcentrifuge tube, and Lactucin-probe or Propargylamine was added (final concentration 100 μM). Both tubes were incubated at RT overnight in a shaker. It was followed by the addition of biotin-azide (Institute of Medicinal Plant Development, Beijing, China) (final concentration 100 μM), then Tris (2-carboxyethyl) phosphine (TCEP, T1656) (final concentration 1 mM), and Tris [(1-benzyl-1H-1,2,3-triazole-4-yl)methyl]-amine (TBTA, T2993) (final concentration 100 μM) (Tokyo Chemical Industry, Japan), and Copper sulfate pentahydrate (CuSO
4·5H
2O) (final concentration 1 mM) (Merck KGaA, Darmstadt, Germany) to each tube with vortex after each addition. Both tubes were incubated at room temperature for 1 h with a vortex every 30 min. Then the tubes were centrifuged at 12,000×
g for 10 min at 4 °C, and the supernatant was removed. Protein precipitate was dissolved by adding 750 µL of pre-cooled methanol and sonicating for 3–4 s at 4 °C using a probe sonicator (~30% power level). It was followed by methanol wash thrice with centrifugation (12,000×
g, 4 °C for 10 min) in between.
4.10. Streptavidin Enrichment
Streptavidin enrichment was by method adopted from the method described by Speers and Cravatt [
35]. Probed protein precipitates were dissolved in PBS using 600 µL of 0.2% SDS. Then 100 µL of streptavidin agarose resin (Pierce™ 20347, Thermo Fisher Scientific, Waltham, MA, USA) was added and mixed for 1 h in a shaker at RT. It is followed by washing with 1 mL of 1× PBS with centrifugation (for 1 min at 2500×
g) in between and each time collection of supernatants. Finally, the streptavidin beads were eluted by adding 100 µL of 2× SDS-loading buffer, boiling in a water bath for 10 min. Cooling at 4 °C and centrifuging at 2500×
g for 1 min. The supernatants were collected and stored at −20 °C until use.
4.11. SDS-PAGE and Staining
Protein samples collected from the previous step were resolved in 10% SDS-PAGE gel and visualized by boiling in 0.25% Coomassie Brilliant Blue (CBB) stain for 5 min [
26]. The blue-stained gel was washed twice by boiling it with distilled water for 5–10 min. Protein bands from sample columns were cut and stored (at −20 °C) until the identification experiment.
4.12. LC-MS/MS Analysis
Resolved protein samples were analyzed using QTRAP® 6500 LC-MS/MS System (AB Sciex LLC, Framingham, MA, USA). Mass spectrometric data were searched against the NCBI database with a taxonomy restriction to 2019 human proteins (172,061 sequences; 53,783,369 residues, 27 July 2020) using MASCOT V2.0 (Matrix Sciences, London, UK).
4.13. Data Analysis
A bar diagram was plotted for the cytotoxicity test to visualize the “mean percent of inhibition ± standard error” of cell proliferation using Microsoft
® Excel 2019 software. The significance of the lactucin doses was compared with control sample data and calculated by “one-way ANOVA” using “IBM
® SPSS
® Statistics 26”. Significant difference at
p 0.05, 0.01, 0.001, and 0.0001 was calculated and expressed with different Asterix “*” numbers on the chart or the data table legend. IC
50 was calculated by plotting the Log10 dose on
X-axis versus the Normalized response on the
Y-axis in Graph Pad Prism 7. Cell cycle cytometry data were analyzed using ModFit LT™ (Version 5.0.9). Apoptosis compensation calculation and data analysis were done using CytExpert™ (Version 2.3.0.84). Protein concentrations were calculated in Microsoft
® Excel 2019. WB bands were quantified using ImageJ
® (Version 1.52a) and analyzed using Graph Pad Prism 7. NMR data was analyzed, the structure was predicted using MestReNova™, and the chemical structure was made using ChemDraw
® Professional. LC-MS/MS results analyzed in MASCOT were combined with WB markers data, and functional enrichment was performed to predict the involved molecular pathway using DAVID Bioinformatics resources [
36,
37].