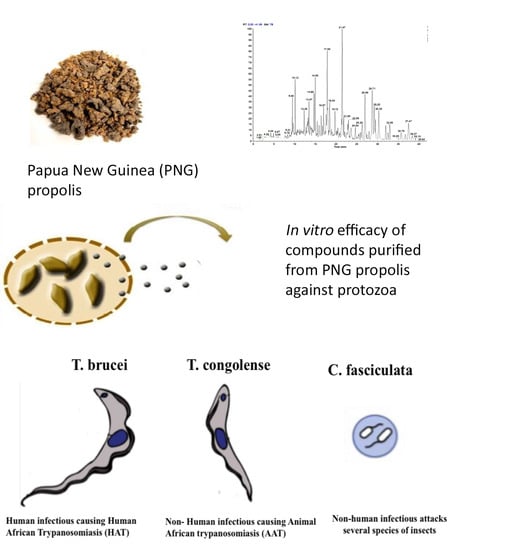
The Antiprotozoal Activity of Papua New Guinea Propolis and Its Triterpenes
Abstract
:1. Introduction
2. Results
2.1. LC-MS Profiling of the Crude PNG Propolis Sample
2.2. Fractionation of the PNG Extract Using Open Column Chromatography (OCC)
2.3. In Vitro Testing of Compounds and Fractions Derived from PNG Propolis against T. b. brucei, T. congolense and C. fasciculata
2.4. Testing of the Cytotoxicity of PNG Extract and Its Fractions against U937 Cells
2.5. Characterisation of MPLC Subfractions Derived from OCC Fractions
2.5.1. Identification of MPLC Fraction PNG-F1-5 as a Mixture of Cycloartenol, 24 (28)-Methylenecycloartenol and Cycloeucalenol
2.5.2. Characterisation of PNG-F4-11 as Betulin
2.5.3. Characterisation of PNG-F4-13 as Betulinic Acid
2.5.4. Characterisation of PNG-F4-18 as Madecassic Acid
2.5.5. Identification of F5 as 20-Hydroxybetulin
2.5.6. Characterisation of PNG-F6-S12 as a Mixture of Mangiferonic Acid and Ambonic Acid
2.5.7. Characterisation of PNG-F6-S15 as a Mixture of Isomangiferolic Acid and Ambolic Acid
2.5.8. Characterisation of PNG-S17 as 27-Hydroxyisomangiferolic Acid
2.6. In Vitro Testing of Compounds of PNG Propolis against T. b. brucei, T. congolense and C. fasciculata
2.6.1. Testing of Compounds and Fractions Derived from PNG Propolis against a Drug-Sensitive (IL3000) and a Diminazene-Resistant Strain (6C3) of T. congolense
2.6.2. Testing of PNG Fractions and Isolated Compounds against U937 Cells
2.6.3. Activity of PNG Propolis and Its Fractions against C. fasciculata
2.6.4. The Effect of 20-Hydroxybetulin on In Vitro Growth of T. b. brucei
3. Discussion
4. Materials and Methods
4.1. General
4.2. Extraction
4.3. Column Chromatography
4.4. Purification
4.5. HPLC–ELSD and LC-MS Analysis
4.6. GC-MS Analysis
4.7. Nuclear Magnetic Resonance Spectroscopy
4.8. Determination of Cytotoxic Effect of PNG Extract and Its Purified Compounds on U937 Mammalian Cells
4.9. Antiprotozoal Assay
4.10. Drug Sensitivity Using Cell Count
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bankova, V.; Popova, M.; Trusheva, B. The phytochemistry of the honeybee. Phytochemistry 2018, 155, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Siheri, W.; Alenezi, S.; Tusiimire, J.; Watson, D.G. The chemical and biological properties of propolis. In Bee Products-Chemical and Biological Properties; Springer: Berlin/Heidelberg, Germany, 2017; pp. 137–178. [Google Scholar]
- Omar, R.M.; Igoli, J.; Gray, A.I.; Ebiloma, G.U.; Clements, C.; Fearnley, J.; Edrada Ebel, R.A.; Zhang, T.; De Koning, H.P.; Watson, D.G. Chemical characterisation of Nigerian red propolis and its biological activity against Trypanosoma brucei. Phytochem. Anal. 2016, 27, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siheri, W.; Zhang, T.; Ebiloma, G.U.; Biddau, M.; Woods, N.; Hussain, M.Y.; Clements, C.J.; Fearnley, J.; Ebel, R.E.; Paget, T.; et al. Chemical and antimicrobial profiling of propolis from different regions within Libya. PLoS ONE 2016, 11, e0155355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siheri, W.; Ebiloma, G.U.; Igoli, J.O.; Gray, A.I.; Biddau, M.; Akrachalanont, P.; Alenezi, S.; Alwashih, M.A.; Edrada-Ebel, R.; Muller, S. Isolation of a novel flavanonol and an alkylresorcinol with highly potent anti-trypanosomal activity from Libyan propolis. Molecules 2019, 24, 1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omar, R.; Igoli, J.O.; Zhang, T.; Gray, A.I.; Ebiloma, G.U.; Clements, C.J.; Fearnley, J.; Ebel, R.E.; Paget, T.; De Koning, H.P. The chemical characterization of Nigerian propolis samples and their activity against Trypanosoma brucei. Sci. Rep. 2017, 7, 923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alotaibi, A.; Ebiloma, G.U.; Williams, R.; Alenezi, S.; Donachie, A.-M.; Guillaume, S.; Igoli, J.O.; Fearnley, J.; De Koning, H.P.; Watson, D.G. European propolis is highly active against trypanosomatids including Crithidia fasciculata. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Cavalcante, G.M.; Camara, C.A.; Silva, E.M.S.D.; Santos, M.S.; Leite, A.B.; Queiroz, A.C.; Evelyn Da Silva, A.; Araújo, M.V.; Alexandre-Moreira, M.S.; Silva, T.M.S. Leishmanicidal Activity of Propolis Collected in the Semiarid Region of Brazil. Front. Pharmacol. 2021, 12, 1625. [Google Scholar] [CrossRef]
- Alotaibi, A.; Ebiloma, G.U.; Williams, R.; Alfayez, I.A.; Natto, M.J.; Alenezi, S.; Siheri, W.; AlQarni, M.; Igoli, J.O.; Fearnley, J.; et al. Activity of compounds from Temperate Propolis against Trypanosoma brucei and Leishmania mexicana. Molecules 2021, 26, 3912. [Google Scholar] [CrossRef]
- Ebiloma, G.U.; Ichoron, N.; Siheri, W.; Watson, D.G.; Igoli, J.O.; De Koning, H.P. The strong anti-kinetoplastid properties of bee propolis: Composition and identification of the active agents and their biochemical targets. Molecules 2020, 25, 5155. [Google Scholar] [CrossRef]
- Giordani, F.; Morrison, L.J.; Rowan, T.G.; De Koning, H.P.; Barrett, M.P. The animal trypanosomiases and their chemotherapy: A review. Parasitology 2016, 143, 1862–1889. [Google Scholar] [CrossRef]
- Simarro, P.P.; Diarra, A.; Ruiz Postigo, J.A.; Franco, J.R.; Jannin, J.G. The human African trypanosomiasis control and surveillance programme of the World Health Organization 2000–2009: The way forward. PLoS Negl. Trop. Dis. 2011, 5, e1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, M.C.; Horn, D.; Fairlamb, A.H.; Ferguson, M.A.; Gray, D.W.; Read, K.D.; De Rycker, M.; Torrie, L.S.; Wyatt, P.G.; Wyllie, S. Anti-trypanosomatid drug discovery: An ongoing challenge and a continuing need. Nat. Rev. Microbiol. 2017, 15, 217–231. [Google Scholar] [CrossRef] [Green Version]
- Stuart, K.; Brun, R.; Croft, S.; Fairlamb, A.; Gürtler, R.E.; McKerrow, J.; Reed, S.; Tarleton, R. Kinetoplastids: Related protozoan pathogens, different diseases. J. Clin. Investig. 2008, 118, 1301–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kipandula, W.; Smith, T.K.; MacNeill, S.A. Tandem affinity purification of exosome and replication factor C complexes from the non-human infectious kinetoplastid parasite Crithidia fasciculata. Mol. Biochem. Parasitol. 2017, 217, 19–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G.; De Graaf, D.C. Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PLoS ONE 2013, 8, e72443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, D.J.; Pilotti, C.A.; Wallis, A.F. Triterpene acids from some Papua New Guinea Terminalia species. Phytochemistry 1992, 31, 881–884. [Google Scholar] [CrossRef]
- Bridges, D.J.; Gould, M.K.; Nerima, B.; Mäser, P.; Burchmore, R.J.; De Koning, H.P. Loss of the high-affinity pentamidine transporter is responsible for high levels of cross-resistance between arsenical and diamidine drugs in African trypanosomes. Mol. Pharmacol. 2007, 71, 1098–1108. [Google Scholar] [CrossRef] [Green Version]
- De Koning, H.P.; Jarvis, S.M. Uptake of pentamidine in Trypanosoma brucei brucei is mediated by the P2 adenosine transporter and at least one novel, unrelated transporter. Acta Trop. 2001, 80, 245–250. [Google Scholar] [CrossRef]
- De Koning, H.P. The drugs of sleeping sickness: Their mechanisms of action and resistance, and a brief history. Trop. Med. Infect. Dis. 2020, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- De, P.T.; Urones, J.; Marcos, I.; Basabe, P.; Cuadrado, M.S.; Moro, R.F. Triterpenes from Euphorbia broteri. Phytochemistry 1987, 26, 1767–1776. [Google Scholar] [CrossRef]
- Hosoe, T.; Nozawa, K.; Udagawa, S.-I.; Nakajima, S.; Kawai, K.-I. An anthraquinone derivative from Dichotomophthora lutea. Phytochemistry 1990, 29, 997–999. [Google Scholar] [CrossRef]
- Tijjani, A.; Ndukwe, I.; Ayo, R. Isolation and characterization of lup-20 (29)-ene-3, 28-diol (Betulin) from the stem-bark of Adenium obesum (Apocynaceae). Trop. J. Pharm. Res. 2012, 11, 259–262. [Google Scholar] [CrossRef] [Green Version]
- Prakash, C.V.S.; Prakash, I. Isolation and structural characterization of Lupane triterpenes from Polypodium vulgare. Res. J. Pharm. Sci. 2012, 1, 23–27. [Google Scholar]
- Matsuda, H.; Morikawa, T.; Ueda, H.; Yoshikawa, M. Medicinal foodstuffs. XXVII. Saponin constituents of gotu kola (2): Structures of new ursane-and oleanane-type triterpene oligoglycosides, centellasaponins B, C, and D, from Centella asiatica cultivated in Sri Lanka. Chem. Pharm. Bull. 2001, 49, 1368–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchino, H.; Satoh, T.; Tanaka, N. Chemical Evaluation of Betula Species in Japan. III. Constituents of Betula maximowicziana. Chem. Pharm. Bull. 1996, 44, 1748–1753. [Google Scholar] [CrossRef] [Green Version]
- Escobedo-Martínez, C.; Concepción Lozada, M.; Hernández-Ortega, S.; Villarreal, M.L.; Gnecco, D.; Enríquez, R.G.; Reynolds, W. 1H and 13C NMR characterization of new cycloartane triterpenes from Mangifera indica. Magn. Reson. Chem. 2012, 50, 52–57. [Google Scholar] [CrossRef]
- Kardar, M.; Zhang, T.; Coxon, G.; Watson, D.; Fearnley, J.; Seidel, V. Characterisation of triterpenes and new phenolic lipids in Cameroonian propolis. Phytochemistry 2014, 106, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Carruthers, L.V.; Munday, J.C.; Ebiloma, G.; Steketee, P.; Jayaraman, S.; Campagnaro, G.D.; Ungogo, M.A.; Lemgruber, L.; Donachie, A.-M.; Rowan, T.G. Diminazene resistance in Trypanosoma congolense is not caused by reduced transport capacity but associated with reduced mitochondrial membrane potential. Mol. Microbiol. 2021, 116, 564–588. [Google Scholar] [CrossRef]
- Munday, J.C.; López, K.E.R.; Eze, A.A.; Delespaux, V.; Van Den Abbeele, J.; Rowan, T.; Barrett, M.P.; Morrison, L.J.; de Koning, H.P. Functional expression of TcoAT1 reveals it to be a P1-type nucleoside transporter with no capacity for diminazene uptake. Int. J. Parasitol. Drugs Drug Resist. 2013, 3, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Pusceddu, M.; Annoscia, D.; Floris, I.; Frizzera, D.; Zanni, V.; Angioni, A.; Satta, A.; Nazzi, F. Honeybees use propolis as a natural pesticide against their major ectoparasite. Proc. R. Soc. B 2021, 288, 20212101. [Google Scholar] [CrossRef]
- Wilson, M.; Brinkman, D.; Spivak, M.; Gardner, G.; Cohen, J. Regional variation in composition and antimicrobial activity of US propolis against Paenibacillus larvae and Ascosphaera apis. J. Invertebr. Pathol. 2015, 124, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Mura, A.; Pusceddu, M.; Theodorou, P.; Angioni, A.; Floris, I.; Paxton, R.J.; Satta, A. Propolis consumption reduces Nosema ceranae infection of European honey bees (Apis mellifera). Insects 2020, 11, 124. [Google Scholar] [CrossRef] [Green Version]
- Habryka, C.; Socha, R.; Juszczak, L. The effect of enriching honey with propolis on the antioxidant activity, sensory characteristics, and quality parameters. Molecules 2020, 25, 1176. [Google Scholar] [CrossRef] [Green Version]
- Saelao, P.; Borba, R.S.; Ricigliano, V.; Spivak, M.; Simone-Finstrom, M. Honeybee microbiome is stabilized in the presence of propolis. Biol. Lett. 2020, 16, 20200003. [Google Scholar] [CrossRef] [PubMed]
- Regan, T.; Barnett, M.W.; Laetsch, D.R.; Bush, S.J.; Wragg, D.; Budge, G.E.; Highet, F.; Dainat, B.; de Miranda, J.R.; Watson, M. Characterisation of the British honey bee metagenome. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravoet, J.; Schwarz, R.S.; Descamps, T.; Yañez, O.; Tozkar, C.O.; Martin-Hernandez, R.; Bartolomé, C.; De Smet, L.; Higes, M.; Wenseleers, T. Differential diagnosis of the honey bee trypanosomatids Crithidia mellificae and Lotmaria passim. J. Invertebr. Pathol. 2015, 130, 21–27. [Google Scholar] [CrossRef]
- Quintana, S.; Plischuk, S.; Brasesco, C.; Revainera, P.; García, M.L.G.; Bravi, M.E.; Reynaldi, F.; Eguaras, M.; Maggi, M. Lotmaria passim (Kinetoplastea: Trypanosomatidae) in honey bees from Argentina. Parasitol. Int. 2021, 81, 102244. [Google Scholar] [CrossRef] [PubMed]
- Bloor, S.J.; Mitchell, K.A. Metabolic products of European-type propolis. Synthesis and analysis of glucuronides and sulfates. J. Ethnopharmacol. 2021, 274, 114035. [Google Scholar] [CrossRef]
- Al Rofaidi, M.; Alotaibi, A.; Aqarni, A.; Alghamdi, A.; Fearnley, J.; Watson, D.G. A Preliminary study of the absorption of and metabolism of temperate propolis by human subjects. J. Food Nutr. Metab. 2020, 3, 1–6. [Google Scholar]
- Shaaban, L.M.S.; Shafeeq, M.A.A.; Mohamed, S.T. Effect of the propolis compared with metronidazole on Giardia lamblia and measurement of Secretory Immunoglobulin A (siga) in intestinal tissue for experimental mice. Plant Arch. 2020, 20, 362–364. [Google Scholar]
- Tran, T.D.; Ogbourne, S.M.; Brooks, P.R.; Sánchez-Cruz, N.; Medina-Franco, J.L.; Quinn, R.J. Lessons from exploring chemical space and chemical diversity of propolis components. Int. J. Mol. Sci. 2020, 21, 4988. [Google Scholar] [CrossRef] [PubMed]
- Yosri, N.; El-Wahed, A.; Aida, A.; Ghonaim, R.; Khattab, O.M.; Sabry, A.; Ibrahim, M.A.; Moustafa, M.F.; Guo, Z.; Zou, X. Anti-Viral and Immunomodulatory Properties of Propolis: Chemical Diversity, Pharmacological Properties, Preclinical and Clinical Applications, and In Silico Potential against SARS-CoV-2. Foods 2021, 10, 1776. [Google Scholar] [CrossRef] [PubMed]
- Popova, M.; Trusheva, B.; Bankova, V. Propolis of stingless bees: A phytochemist’s guide through the jungle of tropical biodiversity. Phytomedicine 2021, 86, 153098. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.; Peyfoon, E.; Zheng, L.; Lu, D.; Seidel, V.; Johnston, B.; Parkinson, J.; Fearnley, J. Application of principal components analysis to 1H-NMR data obtained from propolis samples of different geographical origin. Phytochem. Anal. 2006, 17, 323–331. [Google Scholar] [CrossRef]
- Passmore, J.S.; Lukey, P.T.; Ress, S.R. The human macrophage cell line U937 as an in vitro model for selective evaluation of mycobacterial antigen-specific cytotoxic T-cell function. Immunology 2001, 102, 146–156. [Google Scholar] [CrossRef]
- de Koning, H.P.; MacLeod, A.; Barrett, M.P.; Cover, B.; Jarvis, S.M. Further evidence for a link between melarsoprol resistance and P2 transporter function in African trypanosomes. Mol. Biochem. Parasitol. 2000, 106, 181–185. [Google Scholar] [CrossRef]
- Munday, J.C.; Eze, A.A.; Baker, N.; Glover, L.; Clucas, C.; Aguinaga Andrés, D.; Natto, M.J.; Teka, I.A.; McDonald, J.; Lee, R.S. Trypanosoma brucei aquaglyceroporin 2 is a high-affinity transporter for pentamidine and melaminophenyl arsenic drugs and the main genetic determinant of resistance to these drugs. J. Antimicrob. Chemother. 2014, 69, 651–663. [Google Scholar] [CrossRef]

| RT Min. | [M − H]− | Formula | RDB | Delta ppm | Intensity |
|---|---|---|---|---|---|
| 9.4 | 431.3374 | C24H48O6 | 1.5 | 0.887 | 44.3 × 106 |
| 10.1 | 405.2645 | C24H38O5 | 6.5 | −0.216 | 6.58 × 106 |
| 14.6 | 503.3375 | C30H48O6 | 13 | −0.432 | 4.86 × 106 |
| 14.9 | 487.3425 | C30H48O5 | 13 | −0.744 | 6.31 × 106 |
| 15.8 | 489.3218 | C29H48O6 | 7.5 | −0.659 | 1.03 × 106 |
| 21.4 | 485.3265 | C30H46O5 | 8.5 | −1.396 | 1.12 × 107 |
| 21.8 | 471.3473 | C30H48O4 | 7.5 | −1.280 | 2.18 × 106 |
| 23.0 | 473.3630 453.3373 | C30H50O4 C30H46O3 | 7.0 8.5 | 1.182 −0.237 | 2.02 × 106 1.10 × 106 |
| 25.5 | 469.3318 453.3374 | C30H46O4 C30H47O3 | 8.5 8.5 | −1.029 0.4355 | 6.8 × 105 7.3 × 105 |
| 26.2 | 469.3317 455.3527 | C30H46O4 C30H48O3 | 8.5 7.5 | −1.285 −0.760 | 1.60 × 106 |
| 26.9 | 471.3473 | C30H48O4 | 7.5 | −1.343 | 4.55 × 106 |
| 27.9 | 455.3525 | C30H48O3 | 7.5 | −1.095 | 9.49 × 105 |
| 28.7 | 471.3471 | C30H48O4 | 7.5 | −1.653 | 4.83 × 106 |
| 29.1 | 471.3473 453.3371 | C30H48O4 C30H46O3 | 7.5 8.5 | −1.280 −0.641 | 3.41 × 106 |
| 30.3 | 471.3473 | C30H48O4 | 7.5 | −1.343 | 2.99 × 106 |
| 32.9 | 457.3682 | C30H50O3 | 6.5 | −1.003 | 1.52 × 10 6 |
| 35.6 | 457.3682 | C30H50O3 | 6.5 | −0.937 | 8.5 × 105 |
| 36.7 | 457.3684 | C30H50O3 | 6.5 | −0.544 | 6.63 × 106 |
| 38.3 | 457.3683 | C30H50O3 | 6.5 | −0.740 | 5.33 × 106 |
| Fraction Code | Mass of Fraction (mg) |
|---|---|
| PNG-F1 | 934 |
| PNG-F2 | 301.4 |
| PNG-F3 | 227.2 |
| PNG-F4 | 141.8 |
| PNG-F5 | 54 |
| PNG-F6 | 307.3 |
| PNG-F7 | 172.6 |
| PNG-F8 | 98.3 |
| PNG-F9 | 52.2 |
| PNG-F10 | 32.8 |
| Samples | T. b. brucei S427WT | T. b. brucei B48 | ||||||
|---|---|---|---|---|---|---|---|---|
| EC50 (µg/mL) | EC50 (µg/mL) | |||||||
| AVG | SD | RSD | AVG | SD | RSD | RF | t-Test | |
| PNG crude | 4.0 | 0.095 | 2.40 | 3.85 | 0.37 | 9.52 | 0.96 | 0.50 |
| PNG-F1 | 10.2 | 2.50 | 24.4 | 5.90 | 1.28 | 21.7 | 0.58 | 0.06 |
| PNG-F2 | 14.9 | 1.27 | 8.52 | 9.82 | 2.61 | 26.6 | 0.66 | 0.040 |
| PNG-F3 | 8.2 | 0.73 | 8.93 | 5.82 | 1.38 | 23.6 | 0.71 | 0.05 |
| PNG-F4 | 4.1 | 0.27 | 6.66 | 2.15 | 0.51 | 23.7 | 0.53 | 0.005 |
| PNG-F5 | 2.04 | 0.11 | 5.49 | 2.09 | 0.11 | 11.6 | 0.47 | 0.003 |
| PNG-F6 | 4.8 | 0.60 | 12.4 | 2.13 | 0.50 | 23.5 | 0.44 | 0.004 |
| PNG-F7 | 15.5 | 0.98 | 6.36 | 7.84 | 0.70 | 9.02 | 0.51 | 0.0004 |
| PNG-F8 | 15.8 | 1.40 | 8.87 | 9.16 | 1.76 | 19.2 | 0.58 | 0.007 |
| PNG-F9 | 7.9 | 1.01 | 12.9 | 6.55 | 1.50 | 22.9 | 0.83 | 0.28 |
| PNG-F10 | 15.8 | 1.72 | 9.21 | 12.1 | 2.63 | 21.8 | 0.76 | 0.090 |
| Pentamidine 1 | 0.0034 | 0.0008 | 22.2 | 0.721 | 0.050 | 6.75 | 210 | 0.00001 |
| Samples | AVG EC50 (μg/mL) | ||
|---|---|---|---|
| SD | RSD (%) | ||
| PNG crude | 116.3 | 5.7 | 4.90 |
| PNG-F1 | 83.3 | 11.1 | 13.3 |
| PNG-F2 | 135.1 | 20.1 | 14.9 |
| PNG-F3 | 84.9 | 5.0 | 5.8 |
| PNG-F4 | 40.1 | 1.5 | 3.7 |
| PNG-F5 | 47.0 | 9.63 | 20.5 |
| PNG-F6 | 84.4 | 9.5 | 11.3 |
| PNG-F7 | 57.0 | 6.8 | 11.8 |
| PNG-F8 | 95.6 | 2.3 | 2.4 |
| PNG-F9 | 45.9 | 2.0 | 4.4 |
| PNG-F10 | 53.4 | 5.2 | 9.8 |
| Samples | T. b. brucei S427WT | T. b. brucei B48 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AVG EC50 | AVG EC50 | |||||||||
| μg/mL | μM | SD | RSD | μg/mL | μM | SD | RSD | RF | t-Test | |
| Hydroxybetulin | 2.04 | 4.44 | 0.11 | 5.49 | 0.97 | 2.09 | 0.11 | 11.6 | 0.47 | 0.0003 |
| PNG-F6-S12 | 16.0 | - | 1.47 | 9.21 | 11.3 | - | 2.54 | 22.5 | 0.71 | 0.051 |
| PNG-F6-S15 | 13.5 | - | 0.84 | 6.22 | 12.4 | - | 0.62 | 5.02 | 0.91 | 0.12 |
| Hydroxymangi-ferolic acid | 13.6 | 28.8 | 1.45 | 10.7 | 11.8 | 25.0 | 2.46 | 20.8 | 0.87 | 0.33 |
| PNG-F1-5B | 18.5 | - | 0.72 | 3.90 | 17.6 | - | 0.68 | 3.84 | 0.95 | 0.21 |
| Betulin | 26.6 | 60.1 | 2.69 | 6.26 | 22.3 | 50.5 | 2.39 | 11.8 | 0.84 | 0.13 |
| Betulinic acid | 24.2 | 53.1 | 2.64 | 12.7 | 19.6 | 42.9 | 1.66 | 8.50 | 0.81 | 0.0610 |
| Madecassic acid | 18.1 | 36.0 | 1.81 | 9.45 | 16.0 | 31.7 | 0.91 | 10.4 | 0.88 | 0.23 |
| Pentamidine | - | 0.0043 | 0.0020 | 47.3 | - | 0.62 | 0.10 | 16.5 | 145 | 0.0005 |
| Samples | T. congolense IL3000 | T. congolense 6C3 | ||||||
|---|---|---|---|---|---|---|---|---|
| AVG EC50 | AVG EC50 | |||||||
| μg/mL | SD | RSD | μg/mL | SD | RSD | RF | t-Test | |
| PNG crude | 3.39 | 0.41 | 12.3 | 4.4 | 0.37 | 8.52 | 1.29 | 0.039 |
| PNG-F1 | 13.5 | 3.26 | 24.3 | 13.3 | 3.40 | 25.6 | 0.99 | 0.95 |
| PNG-F2 | 11.4 | 2.49 | 21.8 | 12.0 | 2.34 | 19.5 | 1.05 | 0.79 |
| PNG-F3 | 11.5 | 2.93 | 25.6 | 12.8 | 2.65 | 20.7 | 1.11 | 0.59 |
| PNG-F4 | 10.5 | 2.41 | 23.0 | 9.3 | 0.79 | 8.6 | 0.89 | 0.46 |
| PNG-F5 | 5.77 | 1.28 | 22.2 | 6.09 | 0.33 | 11.8 | 0.48 | 0.017 |
| PNG-F6 | 9.30 | 1.64 | 17.7 | 7.8 | 0.69 | 8.88 | 0.84 | 0.017 |
| PNG-F7 | 12.6 | 1.42 | 11.2 | 12.4 | 2.55 | 20.5 | 0.98 | 0.22 |
| PNG-F8 | 15.6 | 3.10 | 19.9 | 14.6 | 2.64 | 18.1 | 0.93 | 0.90 |
| PNG-F9 | 18.1 | 2.14 | 11.8 | 20.5 | 3.05 | 14.9 | 1.13 | 0.69 |
| PNG-F10 | 18.2 | 3.94 | 21.7 | 21.2 | 4.17 | 19.6 | 1.17 | 0.34 |
| Diminazene 1 | 0.26 | 0.028 | 11.0 | 1.54 | 0.22 | 14.2 | 5.90 | 0.0005 |
| Samples | T. congolense IL3000 | T. congolense 6C3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AVG EC50 | AVG EC50 | |||||||||
| μg/mL | μM | SD | RSD | μg/mL | μM | SD | RSD | RF | t-Test | |
| Hydroxybetulin | 5.77 | 14.4 | 1.28 | 22.2 | 2.77 | 6.09 | 0.33 | 11.8 | 0.48 | 0.017 |
| PNG-F6-S12 | 14.2 | - | 2.75 | 19.4 | 16.9 | - | 4.00 | 23.5 | 1.18 | 0.40 |
| PNG-F6-S15 | 13.7 | - | 2.75 | 20.1 | 13.2 | - | 2.83 | 21.4 | 0.96 | 0.83 |
| Hydroxymangi-ferolic acid | 16.7 | 35.3 | 3.49 | 20.9 | 15.7 | 33.2 | 3.62 | 23.1 | 0.94 | 0.75 |
| PNG-F1-5 | 18.9 | 2.90 | 15.3 | 19.5 | 4.00 | 20.5 | 1.03 | 0.85 | ||
| Betulin | 21.5 | 70.1 | 3.15 | 14.7 | 22.1 | 49.9 | 3.46 | 15.7 | 1.03 | 0.84 |
| Betulinic acid | 17.2 | 44.5 | 2.93 | 17.0 | 18.1 | 28.5 | 3.73 | 20.6 | 1.05 | 0.75 |
| Madecassic acid | 22.6 | 67.6 | 3.25 | 14.4 | 19.8 | 59.1 | 5.37 | 27.1 | 0.88 | 0.48 |
| Diminazene | - | 0.28 | 0.019 | 6.9 | - | 1.55 | 0.22 | 14.2 | 5.5 | 0.0006 |
| Samples | U937 Cells | ||||
|---|---|---|---|---|---|
| AVG EC50 | |||||
| μg/mL | μM | SD | RSD | SI 1 | |
| Hydroxybetulin | 47.0 | 102.0 | 9.63 | 20.50 | 23.0 |
| PNG-F6-S12 | >100 | N/A | 9.55 | 8.92 | >6.3 |
| PNG-F6-S15 | >100 | N/A | 12.30 | 8.44 | >7.4 |
| Hydroxymangiferolic acid | >100 | 260.1 | 7.42 | 6.04 | >7.4 |
| PNGF1-5 | 92.7 | N/A | 6.24 | 6.73 | 5.0 |
| Betulin | 55.8 | 126.0 | 4.34 | 7.78 | 2.1 |
| Betulinic acid | 51.7 | 113.1 | 4.85 | 9.38 | 2.1 |
| Madecassic acid | 90.7 | 179.8 | 5.50 | 6.06 | 5.0 |
| Exp Code | AVG EC50 (μg/mL) | SD | RSD |
|---|---|---|---|
| PNG crude | 20.8 | 1.3 | 6.0 |
| PNG-F1 | 22.1 | 4.44 | 20.1 |
| PNG-F2 | 34.3 | 2.80 | 8.15 |
| PNG-F3 | 28.1 | 1.96 | 7.00 |
| PNG-F4 | 20.8 | 2.73 | 13.1 |
| PNG-F5 | 23.4 | 3.15 | 13.5 |
| PNG-F6 | 53.1 | 4.35 | 8.20 |
| PNG-F7 | 38.5 | 5.67 | 14.7 |
| PNG-F8 | 31.3 | 5.87 | 18.8 |
| PNG-F9 | 24.7 | 1.17 | 4.74 |
| PNG-F10 | 32.2 | 2.29 | 7.10 |
| PAO (µM) | 3.06 | 0.08 | 2.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alenezi, S.S.; Alenezi, N.D.; Ebiloma, G.U.; Natto, M.J.; Ungogo, M.A.; Igoli, J.O.; Ferro, V.A.; Gray, A.I.; Fearnley, J.; de Koning, H.P.; et al. The Antiprotozoal Activity of Papua New Guinea Propolis and Its Triterpenes. Molecules 2022, 27, 1622. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27051622
Alenezi SS, Alenezi ND, Ebiloma GU, Natto MJ, Ungogo MA, Igoli JO, Ferro VA, Gray AI, Fearnley J, de Koning HP, et al. The Antiprotozoal Activity of Papua New Guinea Propolis and Its Triterpenes. Molecules. 2022; 27(5):1622. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27051622
Chicago/Turabian StyleAlenezi, Samya S., Naif D. Alenezi, Godwin U. Ebiloma, Manal J. Natto, Marzuq A. Ungogo, John O. Igoli, Valerie A. Ferro, Alexander I. Gray, James Fearnley, Harry P. de Koning, and et al. 2022. "The Antiprotozoal Activity of Papua New Guinea Propolis and Its Triterpenes" Molecules 27, no. 5: 1622. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27051622