Analysis of Plant–Plant Interactions Reveals the Presence of Potent Antileukemic Compounds
Abstract
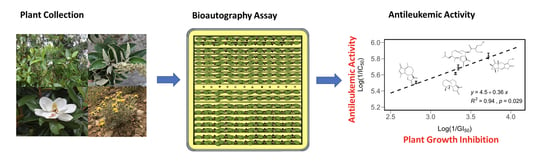
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Plant Materials
3.2. Extraction and Preparation of Extracts
3.3. Bioassay for Germination and Growth Studies
3.4. Contact Bioautography Assay
3.5. Isolation and Characterization of Compounds 1–5
3.6. Antileukemic Assays
3.7. Evaluation of Compounds in Normal Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Mazraati Tajabadi, F.; Pouwer, R.H.; Liu, M.; Dashti, Y.; Campitelli, M.R.; Murtaza, M.; Mellick, G.D.; Wood, S.A.; Jenkins, I.D.; Quinn, R.J. Design and Synthesis of Natural Product Inspired Libraries Based on the Three-Dimensional (3D) Cedrane Scaffold: Toward the Exploration of 3D Biological Space. J. Med. Chem. 2018, 61, 6609–6628. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural Products: A Continuing Source of Novel Drug Leads. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, M.; Williamson, E.M.; Gibbons, S.; Barnes, J.; Prieto-Garcia, J. Fundamentals of Pharmacognosy and Phytotherapy E-BOOK; Elsevier Health Sciences: Amsterdam, The Netherlands, 2017; ISBN 0702070068. [Google Scholar]
- Willis, K.J. State of the World’s Plants Report 2017; Royal Botanic Gardens: London, UK, 2017; ISBN 184246647X. [Google Scholar]
- Fabricant, D.S.; Farnsworth, N.R. The Value of Plants Used in Traditional Medicine for Drug Discovery. Environ. Health Perspect. 2001, 109, 69–75. [Google Scholar]
- Ansari, S. Chapter 24—Overview of Traditional Systems of Medicine in Different Continents. In Preparation of Phytopharmaceuticals for the Management of Disorders; Egbuna, C., Mishra, A.P., Goyal, M.R., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 431–473. ISBN 978-0-12-820284-5. [Google Scholar]
- Tu, Y. Artemisinin—A Gift from Traditional Chinese Medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 10210–10226. [Google Scholar] [CrossRef]
- Achan, J.; Talisuna, A.O.; Erhart, A.; Yeka, A.; Tibenderana, J.K.; Baliraine, F.N.; Rosenthal, P.J.; D’Alessandro, U. Quinine, an Old Anti-Malarial Drug in a Modern World: Role in the Treatment of Malaria. Malar. J. 2011, 10, 144. [Google Scholar] [CrossRef] [Green Version]
- Hartwell, J.L. Plants Used against Cancer: A Survey; Quarterman Publications: Lawrence, MA, USA, 1982; pp. 438–439. [Google Scholar]
- Graham, J.G.; Quinn, M.L.; Fabricant, D.S.; Farnsworth, N.R. Plants Used against Cancer—An Extension of the Work of Jonathan Hartwell. J. Ethnopharmacol. 2000, 73, 347–377. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Plants as a Source of Anti-Cancer Agents. J. Ethnopharmacol. 2005, 100, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Schiffman, J.D.; Fisher, P.G.; Gibbs, P. Early Detection of Cancer: Past, Present, and Future. Am. Soc. Clin. Oncol. Educ. B. 2015, 35, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Sarikaya, I. Breast Cancer and PET Imaging. Nucl. Med. Rev. 2021, 24, 16–26. [Google Scholar] [CrossRef]
- Trinh, A.; Wintermark, M.; Iv, M. Clinical Review of Computed Tomography and MR Perfusion Imaging in Neuro-Oncology. Radiol. Clin. 2021, 59, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Oberlies, N.H.; Kroll, D.J. Camptothecin and Taxol: Historic Achievements in Natural Products Research. J. Nat. Prod. 2004, 67, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Shah, Z.; Gohar, U.F.; Jamshed, I.; Mushtaq, A.; Mukhtar, H.; Zia-UI-Haq, M.; Toma, S.I.; Manea, R.; Moga, M.; Popovici, B. Podophyllotoxin: History, Recent Advances and Future Prospects. Biomolecules 2021, 11, 603. [Google Scholar] [CrossRef] [PubMed]
- Noble, R.L. The Discovery of the Vinca Alkaloids—Chemotherapeutic Agents against Cancer. Biochem. Cell Biol. 1990, 68, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Yao, W. Albumin-Bound Paclitaxel: Worthy of Further Study in Sarcomas. Front. Oncol. 2022, 12, 815900. [Google Scholar] [CrossRef]
- Lahlou, M. Screening of Natural Products for Drug Discovery. Expert Opin. Drug Discov. 2007, 2, 697–705. [Google Scholar] [CrossRef]
- Grkovic, T.; Akee, R.K.; Thornburg, C.C.; Trinh, S.K.; Britt, J.R.; Harris, M.J.; Evans, J.R.; Kang, U.; Ensel, S.; Henrich, C.J.; et al. National Cancer Institute (NCI) Program for Natural Products Discovery: Rapid Isolation and Identification of Biologically Active Natural Products from the NCI Prefractionated Library. ACS Chem. Biol. 2020, 15, 1104–1114. [Google Scholar] [CrossRef] [Green Version]
- Harlev, E.; Nevo, E.; Lansky, E.P.; Lansky, S.; Bishayee, A. Anticancer Attributes of Desert Plants: A Review. Anticancer Drugs 2012, 23, 255–271. [Google Scholar] [CrossRef]
- Keshan, R.; Patra, A.; Mehta, S.; Abdelmotelb, K.F.; Lavale, S.A.; Chaudhary, M.; Aggarwal, S.K.; Chattopadhyay, A. Expression and Regulation of Stress-Responsive Genes in Plants under Harsh Environmental Conditions. In Harsh Environment and Plant Resilience; Springer: Cham, Switzerland, 2021; pp. 25–44. [Google Scholar]
- Hadacek, F. Allelopathy and Chemical Defense. In Chemical Ecology; Hardege, J.D., Ed.; EOLSS Publishers: Oxford, UK, 2009. [Google Scholar]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The Role of Root Exudates in Rhizosphere Interactions with Plants and Other Organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [Green Version]
- Bachheti, A.; Sharma, A.; Bachheti, R.K.; Husen, A.; Pandey, D.P. Plant Allelochemicals and Their Various Applications. In Co-Evolution of Secondary Metabolites; Springer: Cham, Switzerland, 2020; pp. 441–465. [Google Scholar]
- Šoln, K.; Klemenčič, M.; Koce, J.D. Plant Cell Responses to Allelopathy: From Oxidative Stress to Programmed Cell Death. Protoplasma 2022, 1–14. [Google Scholar] [CrossRef]
- Miranda, M.A.F.M.; Varela, R.M.; Torres, A.; Molinillo, J.M.G.; Gualtieri, S.C.J.; Macías, F.A. Phytotoxins from Tithonia Diversifolia. J. Nat. Prod. 2015, 78, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.J. Structure-Activity Relationships of Sesquiterpene Lactones. In Studies in Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 2006; Volume 33, pp. 309–392. ISBN 1572-5995. [Google Scholar]
- Pedrol, N.; González, L.; Reigosa, M.J. Allelopathy and Abiotic Stress. In Allelopathy; Springer: Cham, Switzerland, 2006; pp. 171–209. [Google Scholar]
- Yosef Friedjung, A.; Choudhary, S.P.; Dudai, N.; Rachmilevitch, S. Physiological Conjunction of Allelochemicals and Desert Plants. PLoS ONE 2013, 8, e81580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z. Drought-induced in Vivo Synthesis of Camptothecin in Camptothecaacuminata Seedlings. Physiol. Plant. 2000, 110, 483–488. [Google Scholar] [CrossRef]
- Hoffman, A.; Shock, C.; Feibert, E. Taxane and ABA Production in Yew under Different Soil Water Regimes. HortScience 1999, 34, 882–885. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.-R.; Chang, B.-W.; Zu, Y.-G.; Tang, Z.-H. The Impacts of Increased Nitrate Supply on Catharanthus Roseus Growth and Alkaloid Accumulations under Ultraviolet-B Stress. J. Plant Interact. 2014, 9, 640–646. [Google Scholar] [CrossRef]
- Bamfarahnak, H.; Gholami, A.; Bakzadeh, Z.B.; Hamedi, A.; Mohagheghzadeh, A. Evaluation of Thermal-Stress on the Accumulation of Podophyllotoxin in Shoot in Vitro Cultures of Linum Persicum. Res. J. Pharmacogn. 2014, 1, 3–9. [Google Scholar]
- Majdi, M.; Abdollahi, M.R.; Maroufi, A. Parthenolide Accumulation and Expression of Genes Related to Parthenolide Biosynthesis Affected by Exogenous Application of Methyl Jasmonate and Salicylic Acid in Tanacetum Parthenium. Plant Cell Rep. 2015, 34, 1909–1918. [Google Scholar] [CrossRef]
- Guzman, M.L.; Rossi, R.M.; Karnischky, L.; Li, X.; Peterson, D.R.; Howard, D.S.; Jordan, C.T. The Sesquiterpene Lactone Parthenolide Induces Apoptosis of Human Acute Myelogenous Leukemia Stem and Progenitor Cells. Blood 2005, 105, 4163–4169. [Google Scholar] [CrossRef]
- Tongma, S.; Kobayashi, K.; Usui, K. Allelopathic Activity of Mexican Sunflower [Tithonia Diversifolia (Hemsl.) A. Gray] in Soil under Natural Field Conditions and Different Moisture Conditions. Weed Biol. Manag. 2001, 1, 115–119. [Google Scholar] [CrossRef]
- Wu, T.S.; Shi, L.S.; Kuo, P.C.; Leu, Y.L.; Liou, M.J.; Wu, P.L.; Wu, Y.C.; Iou, S.C.; Chen, Y.P.; Chang, H.C. Cytotoxic Principles from the Leaves of Tithonia Diversifolia. Chin. Pharm. J. 2001, 53, 217–223. [Google Scholar]
- Zhang, A.; Liu, M.; Gu, W.; Chen, Z.; Gu, Y.; Pei, L.; Tian, R. Effect of Drought on Photosynthesis, Total Antioxidant Capacity, Bioactive Component Accumulation, and the Transcriptome of Atractylodes Lancea. BMC Plant Biol. 2021, 21, 293. [Google Scholar] [CrossRef] [PubMed]
- Toscano, S.; Trivellini, A.; Cocetta, G.; Bulgari, R.; Francini, A.; Romano, D.; Ferrante, A. Effect of Preharvest Abiotic Stresses on the Accumulation of Bioactive Compounds in Horticultural Produce. Front. Plant Sci. 2019, 10, 1212. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.-F.; Su, J.; Pan, Z.-H.; Zhang, Z.-J.; Li, X.-N.; Song, L.-D.; Wu, X.-D.; Zhao, Q.-S. Cytotoxic Sesquiterpenoids from the Leaves of Magnolia Grandiflora. Phytochemistry 2018, 155, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, P.E.; Sharma, K.K.; Bystrom, L.M.; Alas, M.A.; Enriquez, R.G.; Malagón, O.; Jones, D.E.; Guzman, M.L.; Compadre, C.M. Dehydroleucodine, a Sesquiterpene Lactone from Gynoxys Verrucosa, Demonstrates Cytotoxic Activity against Human Leukemia Cells. J. Nat. Prod. 2016, 79, 691–696. [Google Scholar] [CrossRef]
- Guamán-Ortiz, L.M.; Bailon-Moscoso, N.; Morocho, V.; Vega-Ojeda, D.; Gordillo, F.; Suárez, A.I. Onoseriolide, from Hedyosmum Racemosum, Induces Cytotoxicity and Apoptosis in Human Colon Cancer Cells. Nat. Prod. Res. 2019, 35, 3151–3155. [Google Scholar] [CrossRef]
- Herz, W.; Govindan, S.V.; Blount, J.F. Glycosidic Disecoeudesmanolides and Other Secosesquiterpene Lactones from Picradeniopsis Species. X-Ray Analysis of Bahia I. J. Org. Chem. 1980, 45, 3163–3172. [Google Scholar] [CrossRef]
- Shui, J.; An, Y.; Ma, Y.; Ichizen, N. Allelopathic Potential of Switchgrass (Panicum virgatum L.) on Perennial Ryegrass (Lolium perenne L.) and Alfalfa (Medicago sativa L.). Environ. Manag. 2010, 46, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, M.; Jefferson, L.V.; Havens, K. Arabidopsis thaliana: A New Test Species for Phytotoxic Bioassays. J. Chem. Ecol. 2005, 31, 1877–1885. [Google Scholar] [CrossRef] [PubMed]
- Ervin, G.N.; Wetzel, R.G. An Ecological Perspective of Allelochemical Interference in Land–Water Interface Communities. Plant Soil 2003, 256, 13–28. [Google Scholar] [CrossRef]
- Rahalison, L.; Hamburger, M.; Hostettmann, K.; Monod, M.; Frenk, E. A Bioautographic Agar Overlay Method for the Detection of Antifungal Compounds from Higher Plants. Phytochem. Anal. 1991, 2, 199–203. [Google Scholar] [CrossRef]
- Suleiman, M.M.; McGaw, L.I.; Naidoo, V.; Eloff, J. Detection of Antimicrobial Compounds by Bioautography of Different Extracts of Leaves of Selected South African Tree Species. Afr. J. Tradit. Complement. Altern. Med. 2010, 7, 64–78. [Google Scholar] [CrossRef]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. Bioautography and Its Scope in the Field of Natural Product Chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Cely-Veloza, W.; Quiroga, D.; Coy-Barrera, E. Quinolizidine-Based Variations and Antifungal Activity of Eight Lupinus Species Grown under Greenhouse Conditions. Molecules 2022, 27, 305. [Google Scholar] [CrossRef]
- Burns, R.M. Silvics of North America; US Department of Agriculture, Forest Service: Washington, DC, USA, 1990; ISBN 0160292603.
- Rathinasabapathi, B.; Ferguson, J.; Gal, M. Evaluation of Allelopathic Potential of Wood Chips for Weed Suppression in Horticultural Production Systems. HortScience 2005, 40, 711–713. [Google Scholar] [CrossRef] [Green Version]
- Abdelgaleil, S.A.M.; Hashinaga, F. Allelopathic Potential of Two Sesquiterpene Lactones from Magnolia grandiflora L. Biochem. Syst. Ecol. 2007, 35, 737–742. [Google Scholar] [CrossRef]
- Hussien, T.A. Chemical Investigation of Secondary Metabolites from Magnolia grandiflora. Sphinx J. Pharm. Med. Sci. 2021, 1, 49–54. [Google Scholar] [CrossRef]
- Marin, G.H.; Mansilla, E. Apoptosis Induced by Magnolia Grandiora Extract in Chlorambucil-Resistant B-Chronic Lymphocytic Leukemia Cells. J. Cancer Res. Ther. 2010, 6, 463–465. [Google Scholar] [CrossRef]
- Carlisi, D.; Lauricella, M.; D’Anneo, A.; de Blasio, A.; Celesia, A.; Pratelli, G.; Notaro, A.; Calvaruso, G.; Giuliano, M.; Emanuele, S. Parthenolide and Its Soluble Analogues: Multitasking Compounds with Antitumor Properties. Biomedicines 2022, 10, 514. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Walsh, E.L.; Ashe, S.; Walsh, J.J. Nature’s Migraine Treatment: Isolation and Structure Elucidation of Parthenolide from Tanacetum parthenium. J. Chem. Educ. 2012, 89, 134–137. [Google Scholar] [CrossRef]
- Siedle, B.; García-Piñeres, A.J.; Murillo, R.; Schulte-Mönting, J.; Castro, V.; Rüngeler, P.; Klaas, C.A.; Da Costa, F.B.; Kisiel, W.; Merfort, I. Quantitative Structure—Activity Relationship of Sesquiterpene Lactones as Inhibitors of the Transcription Factor NF-ΚB. J. Med. Chem. 2004, 47, 6042–6054. [Google Scholar] [CrossRef]
- Ghantous, A.; Gali-Muhtasib, H.; Vuorela, H.; Saliba, N.A.; Darwiche, N. What Made Sesquiterpene Lactones Reach Cancer Clinical Trials? Drug Discov. Today 2010. [Google Scholar] [CrossRef] [PubMed]
- García-Piñeres, A.J.; Castro, V.; Mora, G.; Schmidt, T.J.; Strunck, E.; Pahl, H.L.; Merfort, I. Cysteine 38 in P65/NF-ΚB Plays a Crucial Role in DNA Binding Inhibition by Sesquiterpene Lactones. J. Biol. Chem. 2001, 276, 39713–39720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, A.; Akrap, N.; Marg, B.; Galliardt, H.; Heiligentag, M.; Humpert, F.; Sauer, M.; Kaltschmidt, B.; Kaltschmidt, C.; Seidel, T. Elements of Transcriptional Machinery Are Compatible among Plants and Mammals. PLoS ONE 2013, 8, e53737. [Google Scholar] [CrossRef] [Green Version]
- Ito, M. Conservation and Diversification of Three-Repeat Myb Transcription Factors in Plants. J. Plant Res. 2005, 118, 61–69. [Google Scholar] [CrossRef]
- Roy, S. Function of MYB Domain Transcription Factors in Abiotic Stress and Epigenetic Control of Stress Response in Plant Genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef] [Green Version]
- Walf-Vorderwülbecke, V.; Pearce, K.; Brooks, T.; Hubank, M.; van den Heuvel-Eibrink, M.M.; Zwaan, C.M.; Adams, S.; Edwards, D.; Bartram, J.; Samarasinghe, S. Targeting Acute Myeloid Leukemia by Drug-Induced c-MYB Degradation. Leukemia 2018, 32, 882–889. [Google Scholar] [CrossRef]
- Feng, G.; Burleigh, J.G.; Braun, E.L.; Mei, W.; Barbazuk, W.B. Evolution of the 3R-MYB Gene Family in Plants. Genome Biol. Evol. 2017, 9, 1013–1029. [Google Scholar] [CrossRef]
- Schomburg, C.; Schuehly, W.; Da Costa, F.B.; Klempnauer, K.-H.; Schmidt, T.J. Natural Sesquiterpene Lactones as Inhibitors of Myb-Dependent Gene Expression: Structure–Activity Relationships. Eur. J. Med. Chem. 2013, 63, 313–320. [Google Scholar] [CrossRef]
- Bailly, C. Anticancer Targets and Signaling Pathways Activated by Britannin and Related Pseudoguaianolide Sesquiterpene Lactones. Biomedicines 2021, 9, 1325. [Google Scholar] [CrossRef]
- Singh, A.; Thakur, N.L. Significance of Investigating Allelopathic Interactions of Marine Organisms in the Discovery and Development of Cytotoxic Compounds. Chem. Biol. Interact. 2016, 243, 135–147. [Google Scholar] [CrossRef]
- Baluška, F.; Šamaj, J.; Volkmann, D.; Barlow, P.W. Impact of Taxol-Mediated Stabilization of Microtubules on Nuclear Morphology, Ploidy Levels and Cell Growth in Maize Roots. Biol. Cell 1997, 89, 221–231. [Google Scholar] [CrossRef]
- Vaughan, M.A.; Vaughn, K.C. Mitotic Disrupters from Higher Plants and Their Potential Uses as Herbicides. Weed Technol. 1988, 2, 533–539. [Google Scholar] [CrossRef]
- Fanale, D.; Bronte, G.; Passiglia, F.; Calò, V.; Castiglia, M.; di Piazza, F.; Barraco, N.; Cangemi, A.; Catarella, M.T.; Insalaco, L.; et al. Stabilizing versus Destabilizing the Microtubules: A Double-Edge Sword for an Effective Cancer Treatment Option? Anal. Cell. Pathol. 2015, 2015, 690916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordaliza, M.; Castro, M.; Miguel del Corral, J.; San Feliciano, A. Antitumor Properties of Podophyllotoxin and Related Compounds. Curr. Pharm. Des. 2005, 6, 1811–1839. [Google Scholar] [CrossRef]
- Kamal, A.; Ali Hussaini, S.M.; Rahim, A.; Riyaz, S. Podophyllotoxin Derivatives: A Patent Review (2012–2014). Expert Opin. Ther. Pat. 2015, 25, 1025–1034. [Google Scholar] [CrossRef]
- Oliva, A.; Moraes, R.M.; Watson, S.B.; Duke, S.O.; Dayan, F.E. Aryltetralin Lignans Inhibit Plant Growth by Affecting the Formation of Mitotic Microtubular Organizing Centers. Pestic. Biochem. Physiol. 2002, 72, 45–54. [Google Scholar] [CrossRef]
- Samanta, A.; Maity, T.R.; Das, S.; Datta, A.K.; Datta, S. Effect of Etoposide on Grass Pea DNA Topoisomerase II: An in Silico, in Vivo, and in Vitro Assessments. Bull. Natl. Res. Cent. 2019, 43, 170. [Google Scholar] [CrossRef] [Green Version]
- Buta, J.G.; Kalinski, A. Camptothecin and Other Plant Growth Regulators in Higher Plants with Antitumor Activity. In Biologically Active Natural Products; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1988; Volume 380, pp. 19–294. ISBN 9780841215566. [Google Scholar]
- Sirikantaramas, S.; Yamazaki, M.; Saito, K. Mutations in Topoisomerase I as a Self-Resistance Mechanism Coevolved with the Production of the Anticancer Alkaloid Camptothecin in Plants. Proc. Natl. Acad. Sci. USA 2008, 105, 6782–6786. [Google Scholar] [CrossRef] [Green Version]
- Kantarjian, H.M.; O’Brien, S.; Cortes, J. Homoharringtonine/Omacetaxine Mepesuccinate: The Long and Winding Road to Food and Drug Administration Approval. Clin. Lymphoma Myeloma Leuk. 2013, 13, 530–533. [Google Scholar] [CrossRef] [Green Version]
- Gürel, G.; Blaha, G.; Moore, P.B.; Steitz, T.A. U2504 Determines the Species Specificity of the A-Site Cleft Antibiotics: The Structures of Tiamulin, Homoharringtonine, and Bruceantin Bound to the Ribosome. J. Mol. Biol. 2009, 389, 146–156. [Google Scholar] [CrossRef] [Green Version]
- Prota, A.E.; Bargsten, K.; Diaz, J.F.; Marsh, M.; Cuevas, C.; Liniger, M.; Neuhaus, C.; Andreu, J.M.; Altmann, K.-H.; Steinmetz, M.O. A New Tubulin-Binding Site and Pharmacophore for Microtubule-Destabilizing Anticancer Drugs. Proc. Natl. Acad. Sci. USA 2014, 111, 13817–13821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouëssé, J.; Spielmann, M.; Turpin, F.; Le Chevalier, T.; Azab, M.; Mondésir, J.M. Phase II Study of Elliptinium Acetate Salvage Treatment of Advanced Breast Cancer. Eur. J. Cancer 1993, 29, 856–859. [Google Scholar] [CrossRef]
- Burden, D.A.; Kingma, P.S.; Froelich-Ammon, S.J.; Bjornsti, M.A.; Patchan, M.W.; Thompson, R.B.; Osheroff, N. Topoisomerase II-Etoposide Interactions Direct the Formation of Drug- Induced Enzyme-DNA Cleavage Complexes. J. Biol. Chem. 1996, 271, 29238–29244. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Anticancer Drug | Anticancer Activity | Plant Inhibition Activity |
|---|---|---|
| Paclitaxel | Paclitaxel stabilized microtubes in cancer cells and arrested the replication of cancer cells [16]. | Paclitaxel arrested onion and maize root cells from dividing by stabilizing microtubules [72,73]. |
| Vinblastine | Vinblastine destabilized microtubules in cancer cells and arrested replication [74]. | Vinblastine bound to microtubules and created abnormal multipolar division in Allium cepa L. [73]. |
| Podophyllotoxin, a precursor of etoposide and teniposide | Podophyllotoxin inhibited microtubule organization in cancer cells [75]. Etoposide killed cancer cells by inhibiting topoisomerase II (TopoII) [76]. | Podophyllotoxin inhibited onion (Allium cepa L.) root growth by affecting the formation of mitotic microtubular organizing centers [77]. Etoposide inhibited the division of polyploid cells of grass pea (Lathyrus sativus L.) seedlings. The presumed binding regions of etoposide to TopoII were conserved in plants, Drosophila melanogaster, yeast, and humans [78]. |
| Camptothecin, a precursor to irinotecan and topotecan | Camptothecin killed cancer cells by inhibiting topoisomerase 1 [16]. | Camptothecin selectively caused the inhibition of young developing vascular tissues of the axillary buds of Nicotiana tabacum L. Camptothecin inhibited the sprouting of potatoes by interfering with cell division in the meristem [79]. Early reports showed this using a partially purified enzyme from barley seeds, and strong inhibition of the relaxation of supercoiled pBR322 DNA by the barley DNA enzyme was observed with camptothecin [79]. Later work showed that plants contained a conserved Topo1, and camptothecin-producing plants, including Camptotheca acuminata, Ophiorrhiza pumila, and Ophiorrhiza liukiuensis, had point mutations in Topo1 that conferred resistance to autotoxicity [80]. |
| Homoharringtonine | Homoharringtonine was used for tyrosine kinase inhibitor-resistant chronic myelogenous leukemia (CML). It worked by binding to the A-site of the 80S ribosome and inhibiting translation [81]. | Harringtonine alkaloids, which are related to homoharringtonine, had plant growth regulating activity [79]. The 80S ribosome was conserved across species [82]. |
| Maytansine, a precursor to trastuzumab-emtansine. | Maytansine bound to β-tubulin and blocked the formation of longitudinal tubulin interactions in microtubules [83]. | Maytansine inhibited growth in tobacco callus (Nicotiana tabacum L.) and rice seedling bioassays [79]. |
| Ellipticine, a precursor to elliptinium | Elliptinium is approved in France for the treatment of metastatic breast cancer. Elliptinium and ellipticine inhibited topoisomerase II [84]. | Ellipticine potently inhibited mungbean hypocotyls (Chen, Witham). Ellipticine has been postulated to bind to the same regions of TopoII as etoposide [85]. These regions were conserved in plants, Drosophila melanogaster, yeast, and humans [78]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mery, D.E.; Compadre, A.J.; Ordóñez, P.E.; Selvik, E.J.; Morocho, V.; Contreras, J.; Malagón, O.; Jones, D.E.; Breen, P.J.; Balick, M.J.; et al. Analysis of Plant–Plant Interactions Reveals the Presence of Potent Antileukemic Compounds. Molecules 2022, 27, 2928. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27092928
Mery DE, Compadre AJ, Ordóñez PE, Selvik EJ, Morocho V, Contreras J, Malagón O, Jones DE, Breen PJ, Balick MJ, et al. Analysis of Plant–Plant Interactions Reveals the Presence of Potent Antileukemic Compounds. Molecules. 2022; 27(9):2928. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27092928
Chicago/Turabian StyleMery, David E., Amanda J. Compadre, Paola E. Ordóñez, Edward J. Selvik, Vladimir Morocho, Jorge Contreras, Omar Malagón, Darin E. Jones, Philip J. Breen, Michael J. Balick, and et al. 2022. "Analysis of Plant–Plant Interactions Reveals the Presence of Potent Antileukemic Compounds" Molecules 27, no. 9: 2928. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27092928