3-Amino-5-(indol-3-yl)methylene-4-oxo-2-thioxothiazolidine Derivatives as Antimicrobial Agents: Synthesis, Computational and Biological Evaluation
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Silico Antimicrobial Activity Estimation
2.1.1. Antibacterial Activity
2.1.2. Antifungal Activity
2.1.3. Acute Rat Toxicity
2.2. Chemistry
2.3. Biological Evaluation
2.3.1. Antibacterial Activity
2.3.2. Antifungal Activity
2.3.3. Cytotoxicity Assessment
2.4. Docking Studies
2.4.1. Docking to Antibacterial Targets
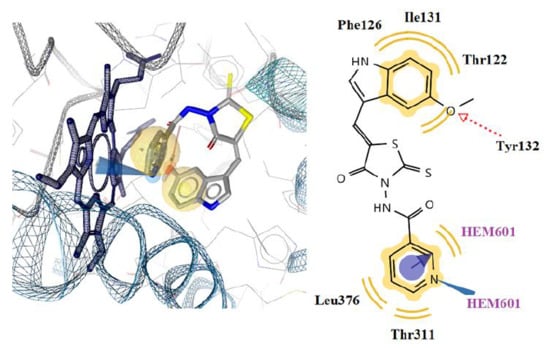
2.4.2. Docking to Lanosterol 14α-demethylase of C. albicans
3. Materials and Methods
3.1. In Silico Biological Activity Evaluation
3.2. Chemistry
3.2.1. General Procedure for the Preparation of N-(4-oxo-2-thioxothiazolidin-3-yl) carbamides 3a–d
3.2.2. General Procedure 5-[(R-1H-indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl] carbamides 5a–k and 5-(R-1H-indol-3-ylmethylene)-3-morpholin-4-yl-2-thioxothiazolidin-4-ones 7a–c
3.3. Antibacterial Activity Evaluation
3.4. Antifungal Evaluation
3.5. Docking Studies
3.6. Cytotoxicity
MTT Assay for Determination of Cell Viability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Michaud, C.M. Global Burden of Infectious Diseases. Encycl. Microbiol. 2009, 444–454. [Google Scholar]
- Nii-Trebi, N.I. Emerging and neglected infectious diseases: Insights, advances, and challenges. Biomed. Res. Int. 2017, 2017, 5245021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S. Emerging Infectious Diseases: Epidemiological Perspective. Indian J. Dermatol. 2017, 62, 459–467. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 16, 145. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Tripathi, A.C.; Gupta, S.J.; Fatima, G.N.; Sonar, P.K.; Verma, A.; Saraf, S.K. 4- Thiazolidinones: The Advances Continue…. Eur. J. Med. Chem. 2014, 7, 52–57. [Google Scholar] [CrossRef]
- Kaminskyy, D.; Kryshchyshyn, A.; Lesyk, R. 5-Ene-4-thiazolidinones–An efficient tool in medicinal chemistry. Eur. J. Med. Chem. 2017, 140, 542–594. [Google Scholar] [CrossRef]
- Kaminskyy, D.; Kryshchyshyn, A.; Lesyk, R. Recent developments with rhodanine as a scaffold for drug discovery. Expert Opin. Drug Discov. 2017, 12, 1233–1252. [Google Scholar]
- Baell, B. Observations on screening-based research and some concerning trends in the literature. Future Med. Chem. 2010, 2, 1529–1546. [Google Scholar]
- Mendgen, T.; Steuer, C.; Klein, C.D. Privileged scaffolds or promiscuous binders: A comparative study on rhodanines and related heterocycles in medicinal chemistry. J. Med. Chem. 2012, 55, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Morphy, R.; Rankovic, Z. Designed multiple ligands. An emerging drug discovery paradigm. J. Med. Chem. 2005, 48, 6523–6543. [Google Scholar] [CrossRef] [PubMed]
- Fortin, S.; Bérubé, G. Advances in the development of hybrid anticancer drugs. Expert Opin. Drug Discov. 2013, 8, 1029–1047. [Google Scholar]
- Kryshchyshyn, A.; Roman, O.; Lozynskyi, A.; Lesyk, R. Thiopyrano[2,3-d]thiazoles as new efficient scaffolds in medicinal chemistry. Sci. Pharm. 2018, 86, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, N.T.; Nhan, H.T.; Van Hung, L.; Thang, T.D.; Kuo, P.C. Synthesis and antibacterial activity of analogs of 5-arylidene-3-(4-methylcoumarin-7-yloxyacetylamino)-2-thioxo-1,3-thiazoli-din-4-one. Molecules 2014, 19, 13577–13586. [Google Scholar] [CrossRef] [Green Version]
- Song, M.-X.; Deng, X.-Q.; Li, Y.-R.; Zheng, C.-J.; Hong, L.; Piao, H.-R. Synthesis and biological evaluation of (E)-1-(substituted)-3-phenylprop-2-en-1-ones bearing rhodanines as potent anti-microbial agents. J. Enzyme Inhib. Med. Chem. 2014, 29, 647–653. [Google Scholar] [CrossRef] [Green Version]
- Krátký, M.; Vinšová, J.; Stolaříková, J. Antimicrobial activity of rhodanine-3-acetic acid derivatives. Bioorg. Med. Chem. 2017, 25, 1839–1845. [Google Scholar] [CrossRef]
- Horishny, V.; Kartsev, V.; Geronikaki, A.; Matiychuk, V.; Petrou, A.; Glamoclija, J.; Ciric, A.; Sokovic, M. 5-(1H-Indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl)alkancarboxylic Acids as Antimicrobial Agents: Synthesis, Biological Evaluation, and Molecular Docking Studies. Molecules 2020, 25, 1964. [Google Scholar] [CrossRef] [Green Version]
- Incerti, M.; Vicini, P.; Geronikaki, A.; Eleftheriou, P.; Tsagkadouras, A.; Zoumpoulakis, P.; Fotakis, C.; Ćirić, A.; Glamočlija, J.; Soković, M. New N-(2-phenyl-4-oxo-1,3-thiazolidin-3-yl)-1,2-benzothiazole -3-carboxamides and Acetamides as Antimicrobial Agents. Med. Chem. Commun. 2017, 8, 2142–2154. [Google Scholar] [CrossRef]
- Ozen, C.; Ceylan-Unlusoy, M.; Aliary, N.; Ozturk, M.; Bozdag-Dundar, O. Thiazolidinedione or Rhodanine: A Study on Synthesis and Anticancer Activity Comparison of Novel Thiazole Derivatives. Pharm. Pharm. Sci. 2017, 20, 415–427. [Google Scholar] [CrossRef]
- Fu, H.; Hou, X.; Wang, L.; Dun, Y.; Yang, X.; Fang, H. Design, synthesis and biological evaluation of 3-aryl-rhodanine benzoic acids as anti-apoptotic protein Bcl-2 inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 5265–5269. [Google Scholar] [CrossRef] [PubMed]
- Havrylyuk, D.; Zimenkovsky, B.; Lesyk, R. Synthesis and Anticancer Activity of Novel Nonfused Bicyclic Thiazolidinone Derivatives. Phosphorus Sulfur 2009, 184, 638–650. [Google Scholar] [CrossRef]
- El-Miligy, M.; Hazzaa, A.; El-Messmary, H.; Nassra, R.A. El-Hawash, Soad, New hybrid molecules combining benzothiophene or benzofuran with rhodanine as dual COX-1/2 and 5-LOX inhibitors: Synthesis, biological evaluation and docking study. Bioorg. Chem. 2017, 72, 102–115. [Google Scholar] [CrossRef] [PubMed]
- R. Atta-Allah, S.; Nassar, I.F.; El-Sayed, W.A. Design, synthesis and anti-inflammatoryvel 5-(Indol-3-yl)-thiazolidinone derivatives as COX-2 inhibitors. J. Pharmacol. Ther. Res. 2020, 5, 1–16. [Google Scholar]
- Powers, J.P.; Piper, D.E.; Li, Y.; Mayorga, V.; Anzola, J.; Chen, J.M.; Jaen, J.C.; Lee, G.; Liu, J.; Peterson, M.G.; et al. SAR and mode of action of novel non-nucleoside inhibitors of hepatitis C NS5b RNA polymerase. J. Med. Chem. 2006, 49, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, K.; Yarovenko, V.N.; Nikitina, A.S.; Zavarzin, I.V.; Krayushkin, M.M.; Kovalenko, L.V.; Esqueda, A.; Odde, S.; Neamati, N. Design, synthesis and structure-activity studies of rhodanine derivatives as HIV-1 integrase inhibitors. Molecules 2010, 15, 3958–3992. [Google Scholar] [CrossRef]
- Petrou, A.; Eleftheriou, P.; Geronikaki, A.; Akrivou, M.G.; Vizirianakis, I. Novel thiazolidin-4-ones as potential non-nucleoside inhibitors of HIV-1 reverse transcriptase. Molecules 2019, 24, 3821. [Google Scholar] [CrossRef] [Green Version]
- Kryshchyshyn, A.; Kaminskyy, D.; Roman, O.; Kralovics, R.; Karpenko, O.; Lesyk, R. Synthesis and anti-leukemic activity of pyrrolidinedione-thiazolidinone hybrids. Ukr Biochem. J. 2020, 92, 108–119. [Google Scholar]
- Havrylyuk, D.; Zimenkovsky, B.; Vasylenko, O.; Gzella, A.; Lesyk, R. Synthesis of new 4- thiazolidinone-, pyrazoline-, and isatin-based conjugates with promising antitumor activity. J. Med. Chem. 2012, 55, 8630–8641. [Google Scholar] [CrossRef]
- Havrylyuk, D.; Roman, O.; Lesyk, R. Synthetic approaches, structure activity relationship and biological applications for pharmacologically attractive pyrazole/pyrazoline–thiazolidine– based hybrids. Eur. J. Med. Chem. 2016, 113, 145–166. [Google Scholar] [CrossRef]
- Saini, T.; Kumar, S.; Narasimhan, B. Central nervous system activities of indole derivatives: An overview. Cent. Nerv. Syst. Agents Med. Chem. 2016, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Singh, O.M. Recent progress in biological activities of indole and indole alkaloids. Mini Rev. Med. Chem. 2018, 18, 9–25. [Google Scholar]
- Kaur, J.; Utreja, D.; Jain, N.; Sharma, S. Recent Developments in the Synthesis and Antimicrobial Activity of Indole and Its Derivatives. Curr. Org. Synth. 2019, 16, 17–37. [Google Scholar] [CrossRef]
- Bathula, C.; Tripathi, S.; Srinivasan, R.; Jha, K.K.; Ganguli, A.; Chakrabarti, G.; Singh, S.; Munshi, P.; Sen, S. Synthesis of novel 5-arylidenethiazolidinones with apoptotic properties via a three component reaction using piperidine as a bifunctional reagent. Org. Biomol. Chem. 2016, 14, 8053–8063. [Google Scholar] [CrossRef]
- Sayed, M.; El-Dean, A.; Ahmed, M.; Hassanien, R. Synthesis of some heterocyclic compounds derived from indole as antimicrobial agents. Synth. Commun. 2018, 48, 413–421. [Google Scholar] [CrossRef]
- Jain, P.; Utreja, D.; Sharma, P. An efficacious synthesis of N-1–, C-3–substituted indole derivatives and their antimicrobial studies. J. Hetrocyclic Chem. 2020, 57, 428–435. [Google Scholar] [CrossRef]
- Shaikh, T.M.A.; Debebe, H. Synthesis and Evaluation of Antimicrobial Activities of Novel N-Substituted Indole Derivatives. J. Chem. 2020, 1–9, Article ID 4358453, 9 pages. [Google Scholar]
- Kumar, P.; Singh, S.; Rizki, M.; Pratama, F. Synthesis of some novel 1H-indole derivatives with antibacterial activity and antifungal activity. Lett. Appl. NanoBioScience 2020, 9, 961–967. [Google Scholar]
- Liu, Z.; Tang, L.; Zhu, H.; Xu, T.; Qiu, C.; Zheng, S.; Gu, Y.; Feng, J.; Zhang, Y.; Liang, G. Design, Synthesis, and Structure−Activity Relationship Study of Novel Indole-2-carboxamide Derivatives as Anti-inflammatoryAgents for the Treatment of Sepsis. J. Med. Chem. 2016, 59, 4637–4650. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Xiao, H.; Bian, Z.; Wang, J. Enantioselective synthesis of indole derivatives byRh/Pd relay catalysis and their anti-inflammatory evaluation. Chem. Commun. 2020, 56, 7573–7576. [Google Scholar] [CrossRef]
- Abdellatif, K.R.A.; Elsaady, M.Y.; Amin, N.H.; Hefny, A.A. Design, Synthesis and biological evaluation of some novel indole derivatives as selective COX-2 inhibitors. J. Appl. Pharm. Sci. 2017, 7, 69–77. [Google Scholar]
- Bhat, M.A.; Al-Omar, M.A.; Raish, M. Indole Derivatives as Cyclooxygenase Inhibitors: Synthesis, Biological Evaluation and Docking Studies. Molecules 2018, 23, 1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidhu, J.S.; Singla, R.; Jaitak, V. Indole Derivatives as Anticancer Agents for Breast Cancer Therapy: A Review. Anticancer Agents Med. Chem. 2015, 16, 160–173. [Google Scholar] [CrossRef] [PubMed]
- El-Sharief, A.M.S.; Ammar, Y.A.; Belal, A.; El-Sharief, M.A.S.; Mohamed, Y.A.; Ahmed, B.M.; Mehany, A.B.M.; Elhag Ali, G.A.M.; Ragab, A. Design, synthesis, molecular docking and biological activity evaluation of some novel indole derivatives as potent anticancer active agents and apoptosis inducers. Bioorg. Chem. 2019, 85, 399–412. [Google Scholar] [CrossRef]
- Cascioferro, S.; Li Petri, G.; Parrino, B.; El Hassouni, B.; Carbone, D.; Arizza, V.; Perricone, U.; Padova, A.; Funel, N.; Peters, G.J.; et al. 3-(6-Phenylimidazo [2,1-b][1,3,4]thiadiazol-2-yl)-1H-Indole Derivatives as New Anticancer Agents in the Treatment of Pancreatic Ductal Adenocarcinoma. Molecules 2020, 25, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.-Z.; Chen, Q.; Yang, G.F. A review on recent developments of indole-containing antiviral agents. J. Med. Chem. 2015, 89, 421–441. [Google Scholar] [CrossRef] [PubMed]
- Bardiot, D. Discovery of Indole Derivatives as Novel and Potent Dengue Virus Inhibitors. J. Med. Chem. 2018, 61, 8390–8401. [Google Scholar] [CrossRef]
- Che, Ζ.; Tian, Υ.; Liu, S.; Hu, M.; Che, G. Discovery of N-arylsulfonyl-3-acylindole benzoyl hydrazone derivatives as anti-HIV-1 agents. Braz. J. Pharm. Sci 2019, 54, e17543. [Google Scholar] [CrossRef] [Green Version]
- Sanna, G.; Madeddu, S.; Giliberti, G.; Piras, S.; Struga, M.; Wrzosek, M.; Kubiak-Tomaszewska, G.; Koziol, A.E.; Savchenko, O.; Lis, T.; et al. Synthesis and Biological Evaluation of Novel Indole-Derived Thioureas. Molecules 2018, 23, 2554. [Google Scholar] [CrossRef] [Green Version]
- Ramya, V.; Vembu, S.; Ariharasivakumar, G.; Gopalakrishnan, M. Synthesis, Characterisation, Molecular Docking, Anti-microbial and Anti-diabetic Screening of Substituted 4-indolylphenyl-6-arylpyrimidine-2-imine Derivatives. Drug Res. (Stuttg) 2017, 67, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Tymiak, A.A.; Rinehart, K.L.; Bakus, G.J. Constituents of morphologically similar sponges: Aplysina and Smenospongia species. Tetrahedron 1985, 41, 1039–1047. [Google Scholar] [CrossRef]
- Djura, P.; Stierle, D.B.; Sullivan, B. Some metabolites of the marine sponges Smenospongia aurea and Smenospongia (ident.Polyfibrospongia) echina. J. Org.Chem. 1980, 45, 1435–1441. [Google Scholar] [CrossRef]
- Fattorusso, E.; Lanzotti, V.; Magno, S.; Novellino, E. Tryptophan derivatives from a Mediterranean anthozoan, Astroides calycularis. J. Nat. Prod. 1985, 48, 924–927. [Google Scholar] [CrossRef]
- Buyukbingol, E.; Suzen, S.; Klopman, G. Studies on the synthesis and structure–activity relationships of 5-(3′-indolyl)-2-thiohydantoin derivatives as aldose reductase enzyme inhibitors. Farmaco 1994, 49, 443–447. [Google Scholar] [PubMed]
- Pogodin, P.V.; Lagunin, A.A.; Rudik, A.V.; Druzhilovskiy, D.S.; Filimonov, D.A.; Poroikov, V.V. AntiBac-Pred: A Web Application for Predicting Antibacterial Activity of Chemical Compounds. J. Chem. Inf. Model. 2019, 59, 4513–4518. [Google Scholar] [CrossRef] [PubMed]
- Poroikov, V.; Filimonov, D.; Gloriozova, T.; Lagunin, A.; Druzhilovskiy, D.; Rudik, A.; Stolbov, L.; Dmitriev, A.; Tarasova, O.; Ivanov, S.; et al. Computer-aided prediction of biological activity spectra for organic compounds: The possibilities and limitations. Russ. Chem. Bull. 2019, 68, 2143–2154. [Google Scholar] [CrossRef]
- Clarivate Analytics Integrity. Available online: https://integrity.clarivate.com/integrity/ (accessed on 21 July 2020).
- Antifungal Activity Predictor. Available online: http://www.way2drug.com/micf (accessed on 21 July 2020).
- Filimonov, D.A.; Zakharov, A.V.; Lagunin, A.A.; Poroikov, V.V. QNA-based ‘Star Track’ QSAR approach. SAR QSAR Env. Res. 2009, 20, 679–709. [Google Scholar] [CrossRef]
- Lagunin, A.; Zakharov, A.; Filimonov, D.; Poroikov, V. QSAR Modelling of Rat Acute Toxicity on the Basis of PASS Prediction. Mol. Inform. 2011, 30, 241–250. [Google Scholar] [CrossRef]
- Bruno, G.; Costantino, L.; Curinga, C.; Maccari, R.; Monforte, F.; Nicolo, F.; Ottana, R.; Vigorita, M.G. Synthesis and Aldose Reductase Inhibitory Activity of5-Arylidene-2,4-thiazolidinediones. Bioorg. Med. Chem. 2002, 10, 1077–1084. [Google Scholar] [CrossRef]
- Abdeen, S.; Kunkle, T.; Salim, N.; Ray, A.-M.; Mammadova, N.; Summers, C.; Stevens, M.; Ambrose, A.J.; Park, Y.; Schultz, P.G.; et al. Sulfonamido-2-arylbenzoxazole GroEL/ES Inhibitors as Potent Antibacterials against Methicillin-Resistant Staphylococcus aureus (MRSA). J. Med. Chem. 2018, 61, 7345–7357. [Google Scholar] [CrossRef]
- Menozzi, G.; Merello, L.; Fossa, P.; Ranise, A.; Mosti, L.; Bondavalli, F.; Loddo, R.; Murgioni, C.; Mascia, V.; La Colla, P.; et al. Synthesis, antimicrobial activity and molecular modeling studies of halogenated 4-[1H-imidazol-1-yl(phenyl)methyl]-1,5-diphenyl-1H-pyrazoles. Bioorg. Med. Chem. 2004, 12, 5465–5483. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.T.; de Lima, G.M.; Maia, J.R.; Speziali, N.L.; Ardisson, J.D.; Rodrigues, L.; Correa, A., Jr.; Romero, O.B. Synthesis, characterization and biocidal activity of new organotin complexes of 2-(3-oxocyclohex-1-enyl)benzoic acid. Eur. J. Med. Chem. 2010, 45, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Gjorgjieva, M.; Tomasicč, T.; Barancokova, M.; Katsamakas, S.; Ilas, J.; Tammela, P.; Masicč, L.P.; Kikelj, D. Discovery of Benzothiazole Scaffold-Based DNA Gyrase B Inhibitors. J. Med. Chem. 2016, 59, 8941–8954. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, N.G.; Manju, P.; Mukesh, M.; Thomas, L. Design, synthesis and molecular docking studies of benzothiazole derivatives as anti microbial agents. Int. J. Pharm. Pharm. Sci. 2013, 5, 101–106. [Google Scholar]
- Ren, Y.; Zhang, L.; Zhou, C.H.; Geng, R.X. Recent Development of Benzotriazole-based Medicinal Drugs. Med. Chem. 2014, 4, 640–662. [Google Scholar] [CrossRef]
- Andres, C.J.; Bronson, J.J.; D’Andrea, S.V.; Walsh, A.W. 4-thiazolidinones: Novel inhibitors of the bacterial enzyme MurB. Bioorg. Med. Chem. Lett. 2000, 10, 715–717. [Google Scholar] [CrossRef]
- Ahmed, S.; Zayed, M.F.; El-Messery, S.M.; Al-Agamy, M.H.; Abdel-Rahman, H.M. Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules 2016, 21, 568. [Google Scholar] [CrossRef]
- Pitta, E.; Tsolaki, E.; Geronikaki, A.; Petrovic, J.; Glamočlija, J.; Sokovic, M.; Crespan, E.; Maga, G.; Bhunia, S.S.; Saxena, A.K. 4-Thiazolidinone derivatives as potent antimicrobial agents: Microwave-assisted synthesis, biological evaluation and docking studies. MedChemComm 2015, 6, 319–326. [Google Scholar] [CrossRef]
- Karanth, S.; Narayana, B.; Kodandoor, S.C.; Sarojini, B.K. 2-{[(4-Hydroxy-3,5-dimethoxyphenyl)methylidene]hydrazinylidene}-4-oxo-1,3-thiazolidin-5-yl Acetic Acid. Molbank 2018, 2018, 2-9 M1009. [Google Scholar] [CrossRef] [Green Version]
- Stana, A.; Vodnar, D.C.; Tamaian, R.; Pîrnău, A.; Vlase, L.; Ionuț, I.; Oniga, O.; Tiperciuc, B. Design, Synthesis and Antifungal Activity Evaluation of New Thiazolin-4-ones as Potential Lanosterol 14α-Demethylase Inhibitors. Int. J. Mol. Sci. 2017, 18, 177. [Google Scholar] [CrossRef]
- Incerti, M.; Vicini, P.; Geronikaki, A.; Eleftheriou, P.; Tsagkadouras, A.; Zoumpoulakis, P.; Fotakis, C.; Ćirić, A.; Glamočlija, J.; Soković, M. New N-(2-phenyl-4-oxo-1,3-thiazolidin-3-yl)-1,2-benzothiazole-3-carboxamides and acetamides asantimicrobial agents. Med. Chem. Commun. 2017, 8, 2142. [Google Scholar] [CrossRef] [PubMed]
- Can, N.O.; Çevik, U.A.; Sağlık, B.N.; Levent, S.; Korkut, B.; Özkay, Y.; Kaplancıklı, Z.A.; Koparal, A.S. Synthesis, Molecular Docking Studies, and Antifungal Activity Evaluation of New Benzimidazole-Triazoles as Potential Lanosterol 14α-Demethylase Inhibitors. J. Chem. 2017, 1–15, Article ID 9387102, 15 pages. [Google Scholar] [CrossRef] [Green Version]
- Benson, T.E.; Walsh, C.T.; Massey, V. Kinetic characterization of wild-type and S229A mutant MurB: Evidence for the role of Ser 229 as a general acid. Biochemistry 1997, 36, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Kartsev, V.; Lichitsky, B.; Geronikaki, A.; Petrou, A.; Smiljkovic, M.; Kostic, M.; Radanovic, O.; Soković, M. Design, synthesis and antimicrobial activity of usnic acid derivatives. MedChemComm 2018, 9, 870–882. [Google Scholar] [CrossRef] [Green Version]
- Fesatidou, M.; Zagaliotis, P.; Camoutsis, C.; Petrou, A.; Eleftheriou, P.; Tratrtat, C.; Haroun, M.; Geronikaki, A.; Ciric, A.; Sokovic, M. 5-Adamantan thiadiazole-based thiazolidinones as antimicrobial agents. Design, synthesis, molecular docking and evaluation. Bioorg. Med. Chem. 2018, 26, 4664–4676. [Google Scholar] [CrossRef]
- Kostić, M.; Smiljković, M.; Petrović, J.; Glamočilija, J.; Barros, L.; Ferreira, I.C.F.R.; Ćirić, A.; Soković, M. Chemical, nutritive composition and a wide range of bioactive properties of honey mushroom Armillaria mellea (Vahl: Fr.) Kummer. Food Function 2017, 8, 3239–3249. [Google Scholar] [CrossRef] [Green Version]
- Kritsi, E.; Matsoukas, M.T.; Potamitis, C.; Detsi, A.; Ivanov, M.; Sokovic, M.; Zoumpoulakis, P. Novel Hit Compounds as Putative Antifungals: The Case of Aspergillus fumigatus. Molecules 2019, 24, 3853. [Google Scholar] [CrossRef] [Green Version]
- Aleksić, M.; Stanisavljević, D.; Smiljković, M.; Vasiljević, P.; Stevanović, M.; Soković, M.; Stojković, D. Pyrimethanil: Between efficient fungicide against Aspergillus rot on cherry tomato and cytotoxic agent on human cell lines. Ann. App. Bio. 2019, 175, 228–235. [Google Scholar] [CrossRef]









| Compound ID | LD50, mg/kg | Toxicity Class | ||||||
|---|---|---|---|---|---|---|---|---|
| IP | IV | Oral | SC | IP | IV | Oral | SC | |
| 5a | 809.8 | 402.5 | 1218 | 780.4 * | 5 | 5 | 4 | 4 * |
| 5b | 680.4 | 309 | 1266 | 1434 | 5 | 5 | 4 | 5 |
| 5c | 980.3 | 311.5 | 1325 | 619.9 * | 5 | 5 | 4 | 4 * |
| 5d | ||||||||
| 5e | 1263 | 466 | 843.9 | 440 * | NT | 5 | 4 | 4 * |
| 5f | 1266 | 502.2 | 469.2 | 477.7 * | NT | 5 | 4 | 4 * |
| 5g | 1010 * | 371.9 | 192.4 * | 397.6 * | 5 * | 5 | 3 * | 4 * |
| 5h | 1282 | 448.5 | 1001 | 1588 | NT | 5 | 4 | 5 |
| 5i | 1299 | 476.5 | 732.3 | 545.4 * | NT | 5 | 4 | 4 * |
| 5j | 1258 | 381.6 | 196.2 * | 422 * | NT | 5 | 3 * | 4 * |
| 5k | 1031 * | 398.3 | 202.8 * | 442 * | 5 * | 5 | 3 * | 4 * |
| 7a | 1033 | 236.6 | 593.7 | 1644 * | 5 | 4 | 4 | 5 * |
| 7b | 1061 | 287.2 | 720 * | 862.1 * | 5 | 4 | 4 * | 4 * |
| 7c | 1180 * | 210.7 | 765.1 * | 682 * | 5 * | 4 | 4 * | 4 * |
| Com/d ID | B.c | M.f | S.a | L.m | En.cl | P.a | S.T | E.coli | |
|---|---|---|---|---|---|---|---|---|---|
| 5a | MIC | 73.3 ± 0.4 | 109.9 ± 0.1 | 73.3 ± 0.3 | 146.5 ± 1.0 | 73.3 ± 0.8 | 73.3 ± 0.08 | 73.3 ± 0.08 | 109.9 ± 0.1 |
| MBC | 146.5 ± 1.0 | 146.5 ± 2.0 | 146.5 ± 2.0 | 293.0 ± 4.0 | 146.5 ± 1.0 | 146.5 ± 1.0 | 146.5 ± 1.0 | 146.5 ± 2.0 | |
| 5b | MIC | 70.5 ± 0.4 | 105.8 ± 0.8 | 70.5 ± 0.8 | 105.8 ± 1.5 | 70.5 ± 0.8 | 70.5 ± 0.8 | 141.0 ± 1.2 | 211.5 ± 2.0 |
| MBC | 141.0 ± 1.0 | 141.0 ± 2.0 | 141.0 ± 1.0 | 141.0 ± 2.0 | 141.0 ± 1.0 | 141.0 ± 0.1 | 282.0 ± 3.0 | 282.0 ± 2.0 | |
| 5c | MIC | 68.2 ± 0.8 | 102.6 ± 1.0 | 68.4 ± 0.5 | 68.4 ± 0.4 | 68.4 ± 0.4 | 68.4 ± 0.4 | 102.6 ± 1.0 | 102.6 ± 1.5 |
| MBC | 136.8 ± 1.0 | 136.8 ± 1.5 | 136.8 ± .1.0 | 136.8 ± 1.0 | 136.8 ± 1.0 | 136.8 ± 1.0 | 136.8 ± 1.5 | 136.8 ± 2.0 | |
| 5d | MIC | 55.6 ± 0.2 | 113.8 ± 0.8 | 37.9 ± 0.2 | 113.8 ± 0.8 | 37.9 ± 0.4 | 37.9 ± 0.2 | 113.8 ± 1.0 | 75.6 ± 0.4 |
| MBC | 75.9 ± 0.4 | 151.7 ± 2.0 | 75.9 ± 0.5 | 151.7 ± 2.0 | 75.9 ± 0.8 | 75.9 ± 0.6 | 151.7 ± 1.0 | 151.7 ± 1.0 | |
| 5e | MIC | 78.9 ± 0.2 | 118.3 ± 1.0 | 57.8 ± 0.4 | 78.9 ± 0.5 | 78.9 ± 0.1 | 78.9 ± 0.6 | 78.9 ± 0.8 | 118.3 ± 1.0 |
| MBC | 157.7 ± 0.8 | 157.7 ± 2.0 | 78.9 ± 0.8 | 157.7 ± 1.5 | 157.7 ± 2.0 | 157.7 ± 1.5 | 157.7 ± 1.0 | 157.7 ± 2.0 | |
| 5f | MIC | 114.1 ± 1.0 | 114.1 ± 8.0 | 114.1 ± 1.0 | 76.1 ± 0.4 | 76.1 ± 0.8 | 76.1 ± 0.3 | 114.1 ± 1.5 | 114.1 ± 1.0 |
| MBC | 152.1 ± 2.0 | 152.1 ± 1.0 | 152.1 ± 1.5 | 152.1 ± 1.0 | 152.1 ± 1.0 | 152.1 ± 1.0 | 152.1 ± 2.0 | 152.1 ± 1.0 | |
| 5g | MIC | 73.1 ± 1.0 | 73.1 ± 1.0 | 53.6 ± 0.4 | 73.1 ± 0.8 | 36.5 ± 0.5 | 109.6 ± 1.0 | 53.6 ± 0.6 | 109.6 ± 1.0 |
| MBC | 146.2 ± 1.0 | 146.2 ± 1.0 | 73.1 ± 0.8 | 146.2 ± 1.6 | 73.1 ± 1.0 | 146.2 ± 1.2 | 73.1 ± 1.0 | 146.2 ± 2.0 | |
| 5h | MIC | 78.9 ± 0.5 | 118.3 ± 1.5 | 118.3 ± 1.0 | 78.9 ± 0.8 | 39.4 ± 0.5 | 39.4 ± 0.6 | 78.9 ± 0.6 | 118.3 ± 1.5 |
| MBC | 157.7 ± 1.0 | 157.7 ± 2.0 | 157.7 ± 2.0 | 157.7 ± 1.0 | 78.9 ± 0.8 | 78.9 ± 0.8 | 157.7 ± 1.2 | 157.7 ± 1.0 | |
| 5i | MIC | 114.1 ± 1.0 | 114.1 ± 1.5 | 76.1 ± 0.8 | 152.1 ± 1.0 | 76.1 ± 0.8 | 76.1 ± 0.8 | 152.1 ± 1.0 | 114.1 ± 1.0 |
| MBC | 152.1 ± 1.0 | 152.1 ± 2.0 | 152.1 ± 2.0 | 304.2 ± 4.0 | 152.1 ± 1.2 | 152.1 ± 1.2 | 304.2 ± 2.0 | 152.1 ± 1.0 | |
| 5j | MIC | 73.1 ± 0.5 | 109.6 ± 1.0 | 58.6 ± 0.4 | 146.2 ± 0.8 | 58.6 ± 0.6 | 58.6 ± 0.8 | 109.6 ± 1.0 | 109.6 ± 1.0 |
| MBC | 146.2 ± 1.0 | 146.2 ± 2.0 | 73.1 ± 0.8 | 292.3 ± 0.2 | 73.1 ± 0.6 | 73.1 ± 0.06 | 146.2 ± 1.0 | 146.2 ± 2.0 | |
| 5k | MIC | 73.1 ± 0.5 | 109.6 ± 1.0 | 58.6 ± 0.4 | 109.6 ± 1.5 | 58.6 ± 0.6 | 58.6 ± 0.6 | 109.6 ± 1.5 | 109.6 ± 2.0 |
| MBC | 146.2 ± 1.0 | 146.2 ± 2.0 | 73.1 ± 0.8 | 146.2 ± 1.0 | 73.1 ± 0.8 | 73.1 ± 0.8 | 146.2 ± 2.0 | 146.2 ± 2.0 | |
| 7a | MIC | 130.3 ± 1.0 | 130.3 ± 1.5 | 63.7 ± 0.4 | 86.9 ± 0.4 | 86.9 ± 1.0 | 86.9 ± 1.0 | 173.7 ± 2.0 | 130.3 ± 2.0 |
| MBC | 173.7 ± 2.0 | 173.7 ± 2.0 | 86.9 ± 0.8 | 173.7 ± 1.5 | 173.7 ± 1.5 | 173.7 ± 1.5 | 347.4 ± 4.0 | 173.7 ± 1.5 | |
| 7b | MIC | 41.7 ± 0.2 | 125.2 ± 1.0 | 41.7 ± 0.2 | 166.9 ± 1.0 | 83.4 ± 0.9 | 61.2 ± 0.5 | 166.9 ± 2.0 | 125.2 ± .1.0 |
| MBC | 83.4 ± 0.4 | 166.9 ± 2.0 | 83.4 ± 0.8 | 333.9 ± .2.0 | 166.9 ± 1.0 | 83.4 ± 1.0 | 333.9 ± 4.0 | 166.9 ± 2.0 | |
| 7c | MIC | 79.9 ± 0.4 | 119.8 ± 1.5 | 79.9 ± 1.0 | 159.8 ± 1.0 | 58.6 ± 0.4 | 58.6 ± 0.4 | 119.8 ± 1.0 | 119.8 ± 1.5 |
| MBC | 159.8 ± 1.0 | 159.8 ± 2.0 | 159.8 ± 1.4 | 319.6 ± 2.0 | 79.9 ± 1.0 | 79.9 ± 1.0 | 159.8 ± 2.0 | 159.8 ± .2.0 | |
| Am. | MIC | 248.0 ± 3.0 | 248.0 ± 2.0 | 248.0 ± 2.0 | 372.0 ± 4.0 | 248.0 ± 3.0 | 744.0 ± 9.0 | 248.0 ± 3.0 | 372.0 ± 4.0 |
| MBC | 372.0 ± 4.0 | 372.0 ± 4.0 | 372.0 ± 2.0 | 744.0 ± 8.0 | 372.0 ± 3.0 | 1240 ± 2 | 492.0 ± 6.0 | 492.0 ± 8.0 | |
| Str. | MIC | 43.0 ± 0.8 | 86.0 ± 1.0 | 172.0 ± 2.0 | 258.0 ± 4.0 | 43.0 ± 0.3 | 172.0 ± 3.0 | 172.0 ± 3.0 | 172.0 ± 2.0 |
| MBC | 86.0 ± 1.0 | 172.0 ± 2.0 | 344.0 ± 4.0 | 516.0 ± 4.0 | 86.0 ± 0.6 | 344.0 ± 3.0 | 344.0 ± 3.0 | 344.0 ± 2.0 |
| Compound ID | Resistant Strains | |||
|---|---|---|---|---|
| MRSA | P.aeruginosa | E.coli | ||
| 5d | MIC | 1260 ± 0.8 | 315 ± 9.0 | 1260 ± 21 |
| MBC | 2520 ± 0.1 | 630 ± 8.0 | 2502 ± 22 | |
| 5g | MIC | 1220 ± 18 | 610 ± 5.0 | 1220 ± 19 |
| MBC | 2440 ± 0.2 | 1202 ± 21 | 2440 ± 16 | |
| 5k | MIC | 1220 ± 0.6 | 610 ± 10 | 1220 ± 0.6 |
| MBC | 2440 ± 22 | 1220 ± 21 | 2440 ± 22 | |
| Streptomycin | MIC | 172.0 ± 21 | 86 ± 12 | 172 ± 21 |
| MBC | - | 172 ± 14 | 344 ± 42 | |
| Ampicilline | MIC | - | 572 ± 64 | 572 ± 78 |
| MBC | / | / | / | |
| Com. ID | A.f | A.v | A.o | A.n | T.v | P.o | P.f | Pvc | |
|---|---|---|---|---|---|---|---|---|---|
| 5a | MIC | 293.0 ± 2.2 | 36.6 ± 0.4 | 26.9 ± 0.1 | 53.7 ± 0.6 | 26.9 ± 0.1 | 36.6 ± 0.2 | 36.6 ± 0.2 | 109.9 ± 0.1 |
| MFC | 586.1 ± 7.0 | 73.3 ± 0.8 | 36.6 ± 0.2 | 73.3 ± 0.8 | 36.6 ± 0.3 | 73.3 ± 0.5 | 73.3 ± 0.8 | 146.5 ± 0.2 | |
| 5b | MIC | 35.2 ± 0.6 | 35.2 ± 0.2 | 25.9 ± 0.2 | 35.2 ± 0.2 | 25.9 ± 0.2 | 35.2 ± 0.5 | 51.7 ± 0.5 | 35.2 ± 0.2 |
| MFC | 70.5 ± 0.6 | 70.5 ± 0.4 | 35.2 ± 0.4 | 70.5 ± 0.8 | 35.2 ± 0.2 | 70.5 ± 0.5 | 70.5 ± 0.5 | 70.5 ± 0.5 | |
| 5c | MIC | 282.0 ± .2.0 | 35.3 ± 0.2 | 18.8 ± 0.2 | 25.9 ± 0.1 | 18.8 ± 0.2 | 35.3 ± 0.2 | 35.3 ± 0.2 | 35.3 ± 0.2 |
| MFC | 564.1 ± 4.0 | 68.2 ± 0.4 | 35.3 ± 0.2 | 35.3 ± 0.2 | 35.3 ± 0.5 | 68.2 ± 0.5 | 68.2 ± 0.5 | 68.2 ± 0.5 | |
| 5d | MIC | 202 ± 0.1 | 37.9 ± 0.2 | 27.8 ± 0.1 | 37.9 ± 0.0 | 27.8 ± 0.2 | 37.9 ± 0.5 | 37.9 ± 0.2 | 37.9 ± 0.2 |
| MFC | 37.9 ± 0.2 | 75.6 ± 0.4 | 37.9 ± 0.5 | 75.6 ± 0.5 | 37.9 ± 0.5 | 75.6 ± 0.5 | 75.6 ± 0.5 | 75.6 ± 0.5 | |
| 5e | MIC | 39.4 ± 0.2 | 39.4 ± 0.2 | 21.0 ± 0.1 | 39.4 ± 0.5 | 21.0 ± 0.1 | 39.4 ± 0.5 | 39.4 ± 0.5 | 39.4 ± 0.5 |
| MFC | 78.9 ± 0.4 | 78.9 ± 0.4 | 39.4 ± 0.2 | 78.9 ± 1.0 | 39.4 ± 0.5 | 78.9 ± 0.5 | 78.9 ± 0.5 | 78.9 ± 0.5 | |
| 5f | MIC | 76.1 ± 0.4 | 38.0 ± 0.2 | 27.9 ± 0.1 | 38.0 ± 0.5 | 27.9 ± 0.2 | 55.8 ± 0.5 | 55.8 ± 0.5 | 55.8 ± 0.5 |
| MFC | 152.1 ± 0.1 | 76.1 ± 0.4 | 38.0 ± 0.5 | 76.1 ± 0.8 | 38.0 ± 0.5 | 76.1 ± 1.0 | 76.1 ± 0.8 | 76.1 ± 1.0 | |
| 5g | MIC | 73.1 ± 0.4 | 19.5 ± 0.2 | 19.5 ± 0.1 | 14.6 ± 0.1 | 9.7 ± 0.01 | 36.5 ± 0.5 | 19.5 ± 0.1 | 26.8 ± 0.1 |
| MFC | 146.2 ± 0.1 | 36.5 ± 0.4 | 36.5 ± 0.5 | 19.5 ± 0.1 | 19.5 ± 0.08 | 73.1 ± 0.8 | 36.5 ± 0.5 | 36.5 ± 0.2 | |
| 5h | MIC | 315.5 ± 2.5 | 78.9 ± 1.0 | 39.4 ± 0.2 | 78.9 ± 0.5 | 28.9 ± 0.1 | 39.4 ± 0.5 | 78.9 ± 0.5 | 118.3 ± 1.0 |
| MFC | 630.9 ± 8.0 | 157.7 ± 1.0 | 78.9 ± 0.8 | 157.7 ± 1.0 | 39.4 ± 0.2 | 78.9 ± 0.5 | 157.7 ± 1.0 | 157.7 ± 2.0 | |
| 5i | MIC | 152.1 ± 1.0 | 38.0 ± 0.4 | 38.0 ± 0.0 | 38.0 ± 0.0 | 27.9 ± 0.2 | 76.1 ± 0.5 | 55.8 ± 0.5 | 76.1 ± 0.5 |
| MFC | 304.2 ± .2.0 | 76.1 ± 0.0 | 76.1 ± 1.0 | 76.1 ± 0.8 | 38.0 ± 0.5 | 152.1 ± 1.0 | 76.1 ± 0.8 | 152.1 ± 1.0 | |
| 5j | MIC | 146.2 ± 1.0 | 35.5 ± 0.2 | 53.6 ± 0.4 | 26.8 ± 0.2 | 35.5 ± 0.5 | 35.5 ± 0.4 | 35.5 ± 0.4 | 35.5 ± 0.6 |
| MFC | 292.3 ± .2.0 | 73.1 ± 0.8 | 73.1 ± 0.8 | 35.5 ± 0.5 | 73.1 ± 1.0 | 73.1 ± 1.0 | 73.1 ± 1.0 | 73.1 ± 1.0 | |
| 5k | MIC | 35.5 ± 0.4 | 35.5 ± 0.2 | 35.5 ± 0.2 | 35.5 ± 0.5 | 35.5 ± 0.2 | 35.5 ± 0.5 | 35.5 ± 0.4 | 35.5 ± 0.2 |
| MFC | 73.1 ± 0.8 | 73.1 ± 0.8 | 73.1 ± 1.0 | 73.1 ± 0.5 | 73.1 ± 0.8 | 73.1 ± 0.8 | 73.1 ± 0.5 | 73.1 ± 0.8 | |
| 7a | MIC | 347.4 ± 2.0 | 43.4 ± 0.2 | 43.4 ± 0.2 | 43.4 ± 0.2 | 31.8 ± 0.2 | 43.4 ± 0.2 | 43.4 ± 0.4 | 86.9 ± 1.0 |
| MFC | 694.8 ± 4.0 | 86.9 ± 1.0 | 86.9 ± 0.5 | 86.9 ± 1.0 | 43.4 ± 0.5 | 86.9 ± 1.0 | 86.9 ± 1.0 | 173.7 ± 2.0 | |
| 7b | MIC | 22.3 ± 0.1 | 22.3 ± 0.1 | 22.3 ± 0.1 | 41.7 ± 0.5 | 22.3 ± 0.2 | 41.7 ± 0.5 | 41.7 ± 1.0 | 22.3 ± 0.1 |
| MFC | 41.7 ± 0.2 | 41.7 ± 0.2 | 41.7 ± 0.5 | 83.4 ± .1.0 | 41.7 ± 0.8 | 83.4 ± 1.0 | 83.4 ± 1.0 | 41.7 ± 0.4 | |
| 7c | MIC | 79.9 ± 0.4 | 21.3 ± 0.1 | 16.0 ± 0.1 | 21.3 ± 0.2 | 10.7 ± 0.2 | 21.3 ± 0.1 | 21.3 ± 1.0 | 58.6 ± 0.4 |
| MFC | 159.8 ± 1.0 | 39.9 ± 0.5 | 21.3 ± 0.1 | 39.9 ± 0.2 | 21.3 ± 0.5 | 39.9 ± 0.2 | 39.9 ± 1.0 | 79.9 ± 1.0 | |
| Ket. | MIC | 380 ± 12 | 2850 ± 68 | 380 ± 12 | 380 ± 8.0 | 475 ± 58 | 3800 ± 58 | 380 ± 16 | 380 ± 12 |
| MFC | 950 ± 23 | 3800 ± 84 | 950 ± 12 | 950 ± 6.0 | 570 ± 86 | 3800 ± 48 | 950 ± 26 | 950 ± 23 | |
| Bif. | MIC | 480 ± 22 | 480 ± .2 | 480 ± 28 | 480 ± 12 | 640 ± 28 | 480 ± 20 | 640 ± 12 | 480 ± 22 |
| MFC | 640 ± 3.4 | 640 ± 0.8 | 800 ± 1.8 | 640 ± 2.3 | 800 ± 3.8 | 640 ± 1.6 | 800 ± 2.1 | 640 ± 3.4 |
| ID | CC50 | B.c | M.f | S.a | L.m | En.cl | P.a | S.T | E.coli | |
|---|---|---|---|---|---|---|---|---|---|---|
| 5d | MIC | 55.6 ± 0.2 | 113.8 ± 0.8 | 37.9 ± 0.2 | 113.8 ± 0.8 | 37.9 ± 0.4 | 37.9 ± 0.2 | 113.8 ± 1.0 | 75.6 ± 0.4 | |
| 252 ± 1.5 | SI | 4.5 | 2.2 | 6.7 | 2.2 | 6.7 | 6.7 | 2.2 | 3.3 | |
| 5g | MIC | 73.1 ± 0.1 | 73.1 ± 1.0 | 53.6 ± 0.4 | 73.1 ± 0.8 | 36.5 ± 0.5 | 109.6 ± 1.0 | 53.6 ± 0.6 | 109.6 ± 1.0 | |
| 256 ± 6.21 | SI | 3.5 | 3.5 | 4.8 | 3.5 | 7.0 | 2.3 | 4.8 | 2.3 | |
| 5k | MIC | 73.1 ± 0.5 | 109.6 ± 1.0 | 58.6 ± 0.4 | 109.6 ± 1.5 | 58.6 ± 0.6 | 58.6 ± 0.6 | 109.6 ± 1.5 | 109.6 ± 2.0 | |
| 252 ± 1.89 | SI | 3.5 | 2.3 | 4.3 | 2.3 | 4.3 | 4.3 | 2.3 | 2.3 | |
| 7c | MIC | 79.9 ± 0.4 | 119.8 ± 1.5 | 79.9 ± 1.0 | 159.8 ± 1.0 | 58.6 ± 0.4 | 58.6 ± 0.4 | 119.8 ± 1.0 | 119.8 ± 1.5 | |
| 225± 1.87 | SI | 2.8 | 1.9 | 2.8 | 1.4 | 3.8 | 3.8 | 1.9 | 1.9 |
| Com. | CC50 (μM) | A.f | A.v | A.o | A.n | T.v | P.o | P.f | Pvc | |
|---|---|---|---|---|---|---|---|---|---|---|
| 5d | MIC | 202 ± 0.1 | 37.9 ± 0.2 | 27.8 ± 0.1 | 37.9 ± 0.2 | 27.8 ± 0.2 | 37.9 ± 0.5 | 37.9 ± 0.2 | 37.9 ± 0.2 | |
| 252 ± 1.5 | SI | 1.3 | 6.7 | 9.1 | 6.7 | 9.1 | 6.7 | 6.7 | 6.7 | |
| 5g | MIC | 73.1 ± 0.2 | 19.5 ± 0.2 | 19.5 ± 0.2 | 14.6 ± 0.1 | 9.7 ± 0.1 | 36.5 ± 0.5 | 19.5 ± 0.1 | 26.8 ± 0.1 | |
| 256 ± 6.21 | SI | 3.5 | 13.1 | 13.1 | 17.5 | 26.4 | 7.0 | 13.1 | 9.6 | |
| 5k | MIC | 35.5 ± 0.4 | 35.5 ± 0.2 | 35.5 ± 0.2 | 35.5 ± 0.5 | 35.5 ± 0.2 | 35.5 ± 0.5 | 35.5 ± 0.4 | 35.5 ± 0.2 | |
| 252 ± 1.89 | SI | 7.1 | 7.1 | 7.1 | 7.1 | 7.1 | 7.1 | 7.1 | 7.1 | |
| 7c | MIC | 79.9 ± 0.4 | 21.3 ± 0.1 | 16.0 ± 0.1 | 21.3 ± 0.2 | 10.7 ± 0.2 | 21.3 ± 0.1 | 21.3 ± 1.0 | 58.6 ± 0.4 | |
| 225 ± 1.87 | SI | 2.8 | 10.6 | 14.1 | 10.6 | 21.0 | 10.6 | 10.6 | 3.8 | |
| Ket. | MIC | 380 ± 12 | 285 ± 68 | 380 ± 12 | 380 ± 8.0 | 475 ± 58 | 380 ± 58 | 380 ± 16 | 380 ± 12 | |
| 60 * | SI | 0.158 | 0.210 | 0.158 | 0.158 | 0.126 | 0.158 | 0.158 | 0.158 |
| Comp. | Est. Binding Energy (kcal/mol) |
I-H E. coli MurB | Residues E. coli MurB | ||
|---|---|---|---|---|---|
|
E.coli DNA Gyrase 1KZN | Thymidylate Kinase 4QGG | E. coli MurB 2Q85 | |||
| 5a | −4.63 | - | −8.22 | 2 | Gly122, Ser228 |
| 5b | −3.12 | - | −7.70 | 2 | Arg213, Asn232 |
| 5c | −5.39 | −2.13 | -9.16 | 2 | Gly122, Ser228 |
| 5d | −6.21 | −4.12 | −10.93 | 2 | Ser228, Arg326 |
| 5e | −6.28 | −2.39 | −8.97 | 2 | Arg213, Ser228 |
| 5f | −5.46 | −1.55 | −8.74 | 2 | Gly122, Ser228 |
| 5g | −6.54 | −3.26 | −10.88 | 3 | Gly122, Ser228, Asn232 |
| 5h | −6.11 | −1.24 | −9.12 | 2 | Gly122, Ser228 |
| 5i | −3.69 | −1.15 | −7.07 | 2 | Arg213, Arg326 |
| 5j | −5.52 | −3.25 | −9.21 | 2 | Gly122, Ser228 |
| 5k | −5.63 | −2.96 | −9.83 | 2 | Arg213, Ser228 |
| 7a | −2.59 | - | −7.28 | 2 | Gly122, Arg213 |
| 7b | −3.67 | - | −7.75 | 2 | Ser228, Asn232 |
| 7c | −4.28 | - | −7.88 | 2 | Gly122, Ser228 |
| N/N | Est. Binding Energy (kcal/mol) CYP51 of C. albicans PDB ID: 5V5Z | I-H |
Residues CYP51 of C. albicans PDB ID: 5V5Z | Interactions with HEM601 |
|---|---|---|---|---|
| 5a | −7.65 | 1 | Tyr132 | Hydrophobic |
| 5b | −9.74 | 1 | Tyr132 | Ionizable, Hydrophobic |
| 5c | −8.13 | 2 | Tyr64, Tyr132 | Hydrophobic |
| 5d | −10.22 | 2 | Tyr118, Tyr132 | Ionizable, Hydrophobic |
| 5e | −9.15 | 1 | Tyr132 | Ionizable, Hydrophobic |
| 5f | −8.79 | 2 | Tyr118, Tyr132 | Hydrophobic |
| 5g | −11.55 | 1 | Tyr132 | Fe binding, Ionizable, Hydrophobic |
| 5h | −7.11 | - | - | Ionizable, Hydrophobic |
| 5i | −7.84 | 1 | Tyr132 | Ionizable, Hydrophobic |
| 5j | −8.72 | 2 | Tyr118, Met508 | Hydrophobic |
| 5k | −9.24 | 2 | Tyr64, Tyr118 | Hydrophobic |
| 7a | −7.08 | 1 | Tyr132 | Hydrophobic |
| 7b | −10.36 | 1 | Tyr132 | Ionizable, Hydrophobic |
| 7c | −10.84 | 2 | Tyr118, Tyr132 | Ionizable, Hydrophobic |
| ketoconazole | −8.23 | 1 | Tyr64 | Ionizable, Hydrophobic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horishny, V.; Kartsev, V.; Matiychuk, V.; Geronikaki, A.; Anthi, P.; Pogodin, P.; Poroikov, V.; Ivanov, M.; Kostic, M.; Soković, M.D.; et al. 3-Amino-5-(indol-3-yl)methylene-4-oxo-2-thioxothiazolidine Derivatives as Antimicrobial Agents: Synthesis, Computational and Biological Evaluation. Pharmaceuticals 2020, 13, 229. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13090229
Horishny V, Kartsev V, Matiychuk V, Geronikaki A, Anthi P, Pogodin P, Poroikov V, Ivanov M, Kostic M, Soković MD, et al. 3-Amino-5-(indol-3-yl)methylene-4-oxo-2-thioxothiazolidine Derivatives as Antimicrobial Agents: Synthesis, Computational and Biological Evaluation. Pharmaceuticals. 2020; 13(9):229. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13090229
Chicago/Turabian StyleHorishny, Volodymyr, Victor Kartsev, Vasyl Matiychuk, Athina Geronikaki, Petrou Anthi, Pavel Pogodin, Vladimir Poroikov, Marija Ivanov, Marina Kostic, Marina D. Soković, and et al. 2020. "3-Amino-5-(indol-3-yl)methylene-4-oxo-2-thioxothiazolidine Derivatives as Antimicrobial Agents: Synthesis, Computational and Biological Evaluation" Pharmaceuticals 13, no. 9: 229. https://0-doi-org.brum.beds.ac.uk/10.3390/ph13090229