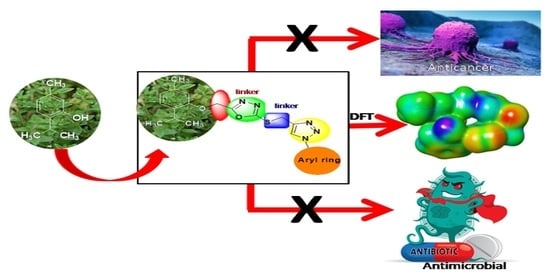
Synthesis and Biological Evaluation of 1,2,3-Triazole Tethered Thymol-1,3,4-Oxadiazole Derivatives as Anticancer and Antimicrobial Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. In Silico Absorption, Distribution, Metabolism and Elimination (ADME)/Pharmacokinetic Studies
2.3. Biological Studies
2.3.1. Antiproliferative Activity
2.3.2. Thymidylate Synthase Activity
2.3.3. Antimicrobial Activity
2.3.4. Computational Studies
Electronic Properties
Molecular Electrostatic and Ionization Potential Maps
2.3.5. Molecular Docking
Thymidylate Synthase (6QXG)
DNA Gyrase B (4URO)
3. Methods and Materials
3.1. Chemistry
3.2. Biological Activities
3.2.1. Antiproliferative Activity
3.2.2. Thymidylate Synthase Activity
3.2.3. Antimicrobial Activity
3.2.4. Statistical Analysis
3.3. Computational Details and Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamal, A.; Bharathi, E.J.; Reddy, J.S.; Ramaiah, M.J.; Dastagiri, D.; Reddy, M.K.; Viswanath, A.; Reddy, T.L.; Shaik, T.B.; Pushpavalli, S.N.C.V.L.; et al. Synthesis and biological evaluation of 3,5-diaryl isoxazoline/isoxazole linked 2,3-dihydroquinazolinone hybrids as anticancer agents. Eur. J. Med. Chem. 2011, 46, 691. [Google Scholar] [CrossRef]
- Parkin, D.M. Global cancer statistics in the year 2000. Lancet Oncol. 2001, 2, 533–543. [Google Scholar] [CrossRef]
- Hassan, M.F.; Rostom, S.A.F.; Badr, M.H.; Ismail, A.E.; Almohammadi, A.M. Synthesis of some 1,4,6-trisubstituted-2-oxo-1,2-dihydropyridine-3-carbonitriles and their biological evaluation as cytotoxic and antimicrobial agents. Arch. Pharm. Chem. Life Sci. 2015, 348, 824–834. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer, the next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.S.; Muzammil, M.H.; Rasool, M.A.; Nisar, R.F.; Alvi, M.A.; Aslam, M.U.; Qamar, M.K.F.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekyere, J.O.; Asante, J. Emerging mechanisms of antimicrobial resistance in bacteria and fungi: Advances in the era of genomics. Future Microbiol. 2018, 13, 241–262. [Google Scholar] [CrossRef] [PubMed]
- Rostom, S.A.F.; Badr, M.H.; Abd-El Razik, H.A.; Ashour, H.M.A.; Abdel-Wahab, A.E. Synthesis of Some Pyrazolines and Pyrimidines Derived from Polymethoxy Chalcones as Anticancer and Antimicrobial Agents. Arch. Pharm. Chem. Life Sci. 2011, 344, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Venkata, S.R.G.; Narkhede, U.C.; Jadhav, V.D.; Naidu, C.G.; Addada, R.R.; Pulya, S.; Ghosh, B. Quinoline Consists of 1H-1,2,3-Triazole Hybrids: Design, Synthesis and Anticancer Evaluation. Chem. Select 2019, 4, 14184–14190. [Google Scholar]
- Ashour, H.F.; Abou-zeid, L.A.; El-Sayed, M.A.A.; Selim, K.A. 1,2,3-Triazole-Chalcone hybrids: Synthesis, in vitro cytotoxic activity and mechanistic investigation of apoptosis induction in multiple myeloma RPMI-8226. Eur. J. Med. Chem. 2020, 189, 112062. [Google Scholar] [CrossRef]
- Lal, K.; Yadav, P.; Kumar, A.; Kumar, A.; Paul, A.K. Design, synthesis, characterization, antimicrobial evaluation and molecular modeling studies of some dehydroacetic acid-chalcone-1,2,3-triazole hybrids. Bioorg. Chem. 2018, 77, 236–244. [Google Scholar] [CrossRef]
- Shaikh, M.H.; Subhedar, D.D.; Nawale, L.; Sarkar, D.; Khan, F.A.K.; Sangshetti, J.N.; Shingate, B.B. 1,2,3-Triazole derivatives as antitubercular agents: Synthesis, biological evaluation and molecular docking study. Med. Chem. Commun. 2015, 6, 1104–1116. [Google Scholar] [CrossRef]
- Kim, T.W.; Yong, Y.; Shin, S.Y.; Jung, H.; Park, K.H.; Lee, Y.H.; Lim, Y.; Jung, K.Y. Synthesis and biological evaluation of phenyl-1H-1, 2, 3-triazole derivatives as anti-inflammatory agents. Bioorg. Chem. 2015, 59, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Brik, A.; Alexandratos, J.; Lin, Y.C.; Elder, J.H.; Olson, A.J.; Wlodawer, A.; Goodsell, D.S.; Wong, C.H. 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. Chem. Biol. Chem. 2005, 6, 1167–1169. [Google Scholar] [CrossRef] [PubMed]
- Gonzaga, D.T.G.; Souza, T.M.L.; Andrade, V.M.M.; Ferreira, V.F.; De C da Silva, F. Identification of 1-aryl-1H-1,2,3-triazoles as potential new antiretroviral agents. Med. Chem. 2018, 14, 242–248. [Google Scholar] [CrossRef]
- Siddiqui, N.; Rana, A.; Khan, S.A.; Haque, S.E.; Alam, M.S.; Ahsan, W.; Ahmed, S. Synthesis of 8-substituted-4-(2/4substitutedphenyl)-2H-[1,3,5]triazino[2,1-b] [1,3]benzo thiazole-2-thiones and their anticonvulsant, antinociceptive, and toxicity evaluation in mice. J. Enz. Inhib. Med. Chem. 2009, 24, 1344–1350. [Google Scholar] [CrossRef]
- Javan, A.J.; Javan, M.J. Electronic structure of some thymol derivatives correlated with the radical scavenging activity: Theoretical study. Food Chem. 2014, 165, 451–459. [Google Scholar] [CrossRef]
- Nesterkina, M.; Kravchenko, I. Synthesis and pharmacological properties of novel esters based on monoterpenoids and glycine. Pharmaceuticals 2017, 10, 47. [Google Scholar] [CrossRef]
- Cui, Z.; Li, X.; Nishida, Y. Synthesis and Bioactivity of Novel Carvacrol and Thymol Derivatives Containing 5-Phenyl-2-furan. Lett. Drug Des. Discov. 2014, 11, 877–885. [Google Scholar] [CrossRef]
- Diab-Assaf, M.; Semaan, J.; El-Sabban, M.; Al-Jouni, S.K.; Azar, R.; Kamal, M.A.; Harakeh, S. Inhibition of Proliferation and Induction of Apoptosis by Thymoquinone via Modulation of TGF Family, p53, p21 and Bcl-2α in Leukemic Cells. Anticancer Agents Med. Chem. 2018, 18, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Malebari, A.M.; Nazreen, S.; Neamatallah, T.; Almalki, A.S.A.; Elhenawy, A.A.; Obaid, R.J.; Alsherif, M.A. Design, synthesis and molecular docking studies of thymol based 1,2,3-triazole hybrids as thymidylate synthase inhibitors and apoptosis inducers against breast cancer cells. Bioorg. Med. Chem. 2021, 38, 116136. [Google Scholar] [CrossRef] [PubMed]
- Rahman, L.; Voeller, D.; Rahman, M.; Lipkowitz, S.; Allegra, C.; Barrett, J.C.; Kaye, F.J.; Zajac-Kaye, M. Thymidylate synthase as an oncogene: A novel role for an essential DNA synthesis enzyme. Cancer Cell 2004, 5, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Catalano, A.; Luciani, R.; Carocci, A.; Cortesi, D.; Pozzi, C.; Borsari, C.; Ferrari, S.; Mangani, S. X-ray crystal structures of Enterococcus faecalis thymidylate synthase with folate binding site inhibitors. Eur. J. Med. Chem. 2016, 123, 649–664. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.M.; Almalki, A.S.A.; Neamatallah, T.; Ali, N.M.; Malebari, A.M.; Nazreen, S. Synthesis of New 1, 3, 4-Oxadiazole-Incorporated 1, 2, 3-Triazole Moieties as Potential Anticancer Agents Targeting Thymidylate Synthase and Their Docking Studies. Pharmaceuticals 2020, 13, 390. [Google Scholar] [CrossRef]
- Lu, G.Q.; Li, X.Y.; Mohamed, O.K.; Wang, D.; Meng, F.H. Design, synthesis and biological evaluation of novel uracil derivatives bearing 1, 2, 3-triazole moiety as thymidylate synthase (TS) inhibitors and as potential antitumor drugs. Eur. J. Med. Chem. 2019, 171, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, D.; Baranowski, D.; Ruszkowski, P.; Boryski, J. Nucleoside dimers analogues with a 1,2,3-triazole linkage: Conjugation of floxuridine and thymidine provides novel tools for cancer treatment. Part II. Nucleosides Nucleotides Nucleic Acids 2019, 38, 807–835. [Google Scholar] [CrossRef]
- Alzhrani, Z.M.M.; Alam, M.M.; Neamatallah, T.; Nazreen, S. Design, synthesis andinvitroantiproliferative activity of new thiazolidinedione-1,3,4-oxadiazole hybrids as thymidylate synthase inhibitors. J. Enzyme Inhib. Med. Chem. 2020, 35, 1116–1123. [Google Scholar] [CrossRef]
- Li, X.; Wang, D.; Lu, G.; Lu, K.; Zhang, T.; Li, S.; Mohamed, K.O.; Xue, W.; Qian, X.; Meng, F. Development of a novel thymidylate synthase (TS) inhibitor capable of up-regulating P53 expression and inhibition angiogenesis in NSCLC. J. Adv. Res. 2020, 26, 95–110. [Google Scholar]
- Du, Q.R.; Li, D.D.; Pi, Y.Z.; Li, J.R.; Sun, J.; Fang, F.; Zhong, W.Q.; Gong, H.B.; Zhu, H.L. Novel 1,3,4-oxadiazole thioether derivatives targeting thymidylate synthase as dual anticancer/antimicrobial agents. Bioorg. Med. Chem. 2013, 21, 2286–2297. [Google Scholar] [CrossRef] [PubMed]
- Nazreen, S. Design, synthesis, and molecular docking studies of thiazolidinediones as PPAR-γ agonists and thymidylate synthase inhibitors. Arch. Der Pharm. 2021, e2100021. [Google Scholar] [CrossRef]
- Balakrishna, K.; Chimbalkar, R.; Prashantha, G. Synthesis and biological activities of some 1,2,4-triazole and 1,3,4-oxadiazoles. Indian J. Het. Chem. 1996, 6, 103–106. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Freener, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Youssef, K.M.; El-Sherbeny, M.A.; El-Shafie, F.S.; Farag, H.A.; Al-Deeb, O.A.; Awadalla, S.A.A. Synthesis of Curcumin Analogues as Potential Antioxidant, Cancer Chemopreventive Agents. Arch. Pharm. 2004, 337, 42–54. [Google Scholar] [CrossRef] [PubMed]
- El Gaafary, M.; Syrovets, T.; Mohamed, H.M.; Elhenawy, A.A.; El-Agrody, A.M.; Amr, A.E.-G.E.; Ghabbour, H.A.; Almehizia, A.A. Synthesis, Cytotoxic Activity, Crystal Structure, DFT Studies and Molecular Docking of 3-Amino-1-(2, 5-dichlorophenyl)-8-methoxy-1H-benzo [f] chromene-2-carbonitrile. Crystals 2021, 11, 184. [Google Scholar] [CrossRef]
- Abusaif, M.S.; Fathy, M.; Abu-Saied, M.A.; Elhenawy, A.A.; Kashyout, A.B.; Selim, M.R.; Ammar, Y.A. New carbazole-based organic dyes with different acceptors for dye-sensitized solar cells: Synthesis, characterization, dssc fabrications and density functional theory studies. J. Mol. Struct. 2021, 1225, 129297. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Thornton, J.M. PROCHECK: Validation of protein-structure coordinates. In International Tables for Crystallography; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Hopkins, A.L.; Keseru, G.M.; Leeson, P.D.; Rees, D.C.; Reynolds, D.H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 2014, 13, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Durcik, M.; Tammela, P.S.M.; Barancokova, M.; Tomasic, T.; Ilas, J.; Kikelj, D.; Zidar, N. Synthesis and Evaluation of N-Phenylpyrrolamides as DNA Gyrase B Inhibitors. Chem. Med. Chem. 2018, 13, 186–198. [Google Scholar] [CrossRef] [Green Version]
- Alghamdi, A.A.; Alam, M.M.; Nazreen, S. In silico ADME predictions and in vitro antibacterial evaluation of 2-hydroxy benzothiazole-based 1,3,4-oxadiazole derivatives. Turk. J. Chem. 2020, 44, 1068–1084. [Google Scholar] [CrossRef]








| No. | Lipinski Parameters | nROTB e | TPSA f | %ABS g | BBB h | GI ABS i | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MW a | HBA b | HBD c | iLogP d | Violations | ||||||
| 5 | 302.39 | 4 | 0 | 3.61 | 0 | 6 | 73.45 | 83.65 | Yes | High |
| 6 | 421.52 | 6 | 0 | 4.14 | 0 | 8 | 104.16 | 73.07 | No | High |
| 7 | 451.54 | 7 | 0 | 4.54 | 1 | 9 | 113.39 | 69.88 | No | High |
| 8 | 465.52 | 8 | 1 | 3.93 | 0 | 9 | 141.46 | 60.20 | No | Low |
| 9 | 437.51 | 7 | 1 | 4.08 | 0 | 8 | 124.39 | 66.09 | No | Low |
| 10 | 455.96 | 6 | 0 | 4.22 | 0 | 8 | 104.16 | 73.07 | No | High |
| 11 | 455.96 | 6 | 0 | 4.05 | 0 | 8 | 104.16 | 73.07 | No | High |
| 12 | 490.41 | 6 | 0 | 4.45 | 1 | 8 | 104.16 | 73.07 | No | Low |
| 13 | 500.41 | 6 | 0 | 4.61 | 1 | 8 | 104.16 | 73.07 | No | High |
| 14 | 500.41 | 6 | 0 | 4.19 | 1 | 8 | 107.16 | 73.07 | No | High |
| 15 | 439.51 | 7 | 0 | 4.29 | 0 | 8 | 104.16 | 73.07 | No | High |
| 16 | 457.50 | 8 | 0 | 4.30 | 1 | 8 | 104.16 | 73.07 | No | Low |
| 17 | 492.59 | 7 | 1 | 3.26 | 0 | 11 | 133.26 | 63.02 | No | Low |
| 18 | 532.41 | 8 | 0 | 3.99 | 1 | 8 | 121.38 | 67.13 | No | Low |
| Compound | MCF-7 | HCT-116 | HepG2 |
|---|---|---|---|
| 6 | 22.5 | 36.5 | 41.6 |
| 7 | 2.4 | 3.1 | 1.8 |
| 8 | 3.9 | 5.8 | 5.3 |
| 9 | 1.1 | 2.6 | 1.4 |
| 10 | 1.3 | 3.8 | 2.5 |
| 11 | 4.9 | 16.3 | 18.6 |
| 12 | 59.4 | 47.3 | 40.6 |
| 13 | 65.3 | 78.7 | 69.4 |
| 14 | 41.8 | 32.8 | 23.7 |
| 15 | 23.15 | 31.4 | 49.2 |
| 16 | 34.5 | 56.2 | 58.3 |
| 17 | 9.39 | 17.3 | 39.1 |
| 18 | 98.2 | 118.5 | 95.7 |
| Dox | 1.2 | 2.5 | 1.8 |
| 5-FU | 18.74 | 30.68 | 28.65 |
| Compounds | IC50 (µM) |
|---|---|
| 7 | 3.52 ± 1.2 |
| 8 | 3.98 ± 1.1 |
| 9 | 1.95 ± 0.9 |
| 10 | 2.18 ± 1.7 |
| 11 | 4.24 ± 0.8 |
| Pemetrexed | 7.26 ± 1.1 |
| Compound No. | E. coli | S. aureus | C. albicans |
|---|---|---|---|
| 5 | 12 ± 0.32 | 9 ± 0.30 | 10 ± 0.32 |
| 6 | 14 ± 0.38 | 10 ± 0.40 | --- |
| 7 | 12 ± 0.34 | 12 ± 0.34 | --- |
| 8 | --- | 10 ± 0.40 | --- |
| 9 | 12 ± 0.36 | 12 ± 0.34 | 10 ± 0.032 |
| 10 | --- | --- | --- |
| 11 | --- | --- | --- |
| 12 | 10 ± 0.38 | --- | 12 ± 0.036 |
| 13 | 11 ± 0.34 | --- | --- |
| 14 | 9 ± 0.32 | --- | --- |
| 15 | --- | --- | --- |
| 16 | 14 ± 0.34 | 15±0.38 | --- |
| 17 | 12 ± 0.32 | 12 ± 0.34 | --- |
| 18 | --- | --- | --- |
| Novobiocin | 14 ± 0.64 | 10 ± 0.24 | NT |
| Fluconazole | NT | NT | 16 ± 0.60 |
| Compd. | E HOMO | E LUMO | ΔE HOMO/LUMO | E HOMO-1 | E LUMO+1 | ΔE HOMO-1/LUMO+1 | IP | η | S | χ | ω |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | −6.026 | −1.802 | −4.224 | −0.150 | −0.062 | −0.0881 | 5.83 | 2.612 | 0.383 | 2.231 | −3.414 |
| 6 | −5.834 | −1.472 | −4.362 | −1.469 | −0.707 | −0.7619 | 5.72 | 2.181 | 0.458 | 3.059 | −3.653 |
| 7 | −5.995 | −1.200 | −4.795 | −1.360 | −1.159 | −0.215 | 1.87 | 2.398 | 0.417 | 2.699 | −3.597 |
| 8 | −5.836 | −1.987 | −3.849 | −1.361 | −0.424 | −0.9361 | 5.71 | 1.924 | 0.520 | 3.976 | −3.912 |
| 9 | −6.104 | −1.227 | −4.877 | −0.963 | −0.435 | −0.5279 | 5.82 | 2.439 | 0.410 | 2.755 | −3.666 |
| 10 | −6.139 | −1.368 | −4.771 | −0.931 | −0.844 | −0.0871 | 6.11 | 2.386 | 0.419 | 2.952 | −3.753 |
| 11 | −5.904 | −1.794 | −4.110 | −1.083 | −0.615 | −0.4680 | 5.69 | 2.055 | 0.487 | 3.604 | −3.849 |
| 12 | −6.181 | −1.588 | −4.593 | −1.197 | −0.990 | −0.2068 | 5.67 | 2.296 | 0.435 | 3.285 | −3.884 |
| 13 | −5.861 | −1.739 | −4.122 | −1.069 | −0.459 | −0.6106 | 5.67 | 2.061 | 0.485 | 3.502 | −3.800 |
| 14 | −5.892 | −1.828 | −4.064 | −1.140 | −0.552 | −0.5875 | 5.90 | 2.032 | 0.492 | 3.666 | −3.860 |
| 15 | −5.849 | −1.516 | −4.332 | −1.026 | −0.422 | −0.6041 | 5.77 | 2.166 | 0.462 | 3.130 | −3.683 |
| 16 | −6.218 | −1.473 | −4.745 | −1.031 | −1.004 | −0.0272 | 6.07 | 2.372 | 0.422 | 3.117 | −3.846 |
| 17 | −5.961 | −1.277 | −4.684 | −0.917 | −0.705 | −0.2122 | 5.92 | 2.342 | 0.427 | 2.796 | −3.619 |
| 18 | −5.989 | −1.821 | −4.168 | −1.625 | −1.059 | −0.5660 | 5.70 | 2.084 | 0.480 | 3.659 | −3.905 |
| Cpd. | ΔE | RMSD | Eplace | Econf | Eele | LE | Ki | LEscale | FQ |
|---|---|---|---|---|---|---|---|---|---|
| FdUMP | −7.90 | 0.94 | −430.17 | −15.64 | −14.74 | −8.39 | 2.47 | 3.30 | 5.09 |
| 5FU | −5.28 | 1.60 | −433.91 | −21.32 | −17.47 | −3.30 | 2.41 | −5.28 | −5.28 |
| 5 | −6.42 | 1.26 | 2.48 | −14.45 | −9.31 | −5.08 | 1.98 | 3.05 | 2.03 |
| 6 | −7.36 | 1.64 | 41.66 | −17.27 | −10.34 | −4.48 | 1.54 | 2.82 | 1.67 |
| 7 | −7.50 | 1.10 | 39.34 | −17.66 | −8.68 | −6.81 | 2.16 | 3.17 | 3.64 |
| 8 | −7.59 | 1.92 | 19.80 | −7.35 | −7.87 | −3.95 | 1.73 | 2.68 | 1.27 |
| 9 | −7.46 | 1.63 | 32.10 | −8.59 | −9.55 | −4.56 | 1.94 | 2.82 | 1.74 |
| 10 | −7.14 | 1.01 | 20.09 | −14.91 | −7.02 | −7.04 | 1.56 | 3.24 | 3.80 |
| 11 | −7.04 | 1.53 | 36.13 | −14.00 | −7.68 | −4.61 | 0.16 | 2.88 | 1.73 |
| 12 | −6.87 | 1.73 | 35.84 | −18.02 | −9.06 | −3.97 | 1.81 | 2.77 | 1.20 |
| 13 | −7.39 | 1.40 | 48.40 | −17.34 | −12.24 | −5.27 | 1.43 | 2.96 | 2.32 |
| 14 | −7.50 | 1.33 | 40.76 | −12.81 | −8.17 | −5.63 | 1.49 | 3.00 | 2.63 |
| 15 | −7.04 | 1.46 | 43.98 | −23.03 | −10.03 | −4.82 | 0.94 | 2.92 | 1.90 |
| 16 | −7.17 | 1.65 | 32.20 | −10.86 | −9.15 | −4.36 | 1.88 | 2.81 | 1.54 |
| 17 | −8.16 | 1.60 | −4.51 | −17.16 | −8.75 | −5.10 | 2.03 | 2.84 | 2.26 |
| 18 | −7.45 | 1.56 | 131.24 | −15.92 | −10.20 | −4.78 | 1.77 | 2.86 | 1.91 |
| Cpd. | ΔE | RMSD | Eplace | Econf | Eele | LE | Ki | LEscale | FQ |
|---|---|---|---|---|---|---|---|---|---|
| Nov. | −8.41 | 1.16 | 10.98 | −9.99 | −7.80 | −7.24 | 1.95 | 3.56 | 8.26 |
| 5 | −9.18 | 1.97 | 48.83 | −10.04 | −8.81 | −4.67 | 1.86 | 3.12 | 4.12 |
| 6 | −9.57 | 1.11 | 57.74 | −5.82 | −2.67 | −8.65 | 1.82 | 2.66 | 2.01 |
| 7 | −8.95 | 1.58 | 28.83 | −12.31 | −12.87 | −5.66 | 1.89 | 3.16 | 5.49 |
| 8 | −8.26 | 1.19 | 32.72 | −5.94 | −6.46 | −6.93 | 1.97 | 2.85 | 2.81 |
| 9 | −9.26 | 1.94 | 31.38 | −9.99 | −9.03 | −4.77 | 1.86 | 3.10 | 3.83 |
| 10 | −6.09 | 1.79 | 31.82 | −43.28 | −2.51 | −1.17 | 3.34 | 2.67 | 2.10 |
| 11 | −8.49 | 1.38 | 60.48 | −6.87 | −1.93 | −6.14 | 1.94 | 2.74 | 1.57 |
| 12 | −8.21 | 1.96 | 48.99 | −6.93 | −6.42 | −4.19 | 1.98 | 2.97 | 3.17 |
| 13 | −8.26 | 1.87 | 41.56 | −9.63 | −7.28 | −4.42 | 1.97 | 2.66 | 1.52 |
| 14 | −6.09 | 0.82 | 41.09 | −41.70 | −2.56 | −2.56 | 1.96 | 2.70 | 1.72 |
| 15 | −8.08 | 1.23 | 30.40 | −8.24 | −6.28 | −6.58 | 1.99 | 3.43 | 0.86 |
| 16 | −9.62 | 1.26 | 14.00 | −5.65 | −9.83 | −7.61 | 1.82 | 3.07 | 3.51 |
| 17 | −8.68 | 1.48 | 129.68 | −7.59 | −10.45 | −5.85 | 1.92 | 3.05 | 4.56 |
| 18 | −8.26 | 0.70 | 44.95 | −22.82 | −12.90 | −11.83 | 1.97 | 2.91 | 2.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almalki, A.S.A.; Nazreen, S.; Malebari, A.M.; Ali, N.M.; Elhenawy, A.A.; Alghamdi, A.A.A.; Ahmad, A.; Alfaifi, S.Y.M.; Alsharif, M.A.; Alam, M.M. Synthesis and Biological Evaluation of 1,2,3-Triazole Tethered Thymol-1,3,4-Oxadiazole Derivatives as Anticancer and Antimicrobial Agents. Pharmaceuticals 2021, 14, 866. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14090866
Almalki ASA, Nazreen S, Malebari AM, Ali NM, Elhenawy AA, Alghamdi AAA, Ahmad A, Alfaifi SYM, Alsharif MA, Alam MM. Synthesis and Biological Evaluation of 1,2,3-Triazole Tethered Thymol-1,3,4-Oxadiazole Derivatives as Anticancer and Antimicrobial Agents. Pharmaceuticals. 2021; 14(9):866. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14090866
Chicago/Turabian StyleAlmalki, Abdulraheem S. A., Syed Nazreen, Azizah M. Malebari, Nada M. Ali, Ahmed A. Elhenawy, Abdullah A. A. Alghamdi, Abrar Ahmad, Sulaiman Y. M. Alfaifi, Meshari A. Alsharif, and Mohammad Mahboob Alam. 2021. "Synthesis and Biological Evaluation of 1,2,3-Triazole Tethered Thymol-1,3,4-Oxadiazole Derivatives as Anticancer and Antimicrobial Agents" Pharmaceuticals 14, no. 9: 866. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14090866