Impact of the Uridine–Cytidine Kinase Like-1 Protein and IL28B rs12979860 and rs8099917 SNPs on the Development of Hepatocellular Carcinoma in Cirrhotic Chronic Hepatitis C Patients—A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Analysis of UCKL-1 Expression in the Liver Tissue
2.3. Detection of Single-Nucleotide Polymorphism in IL-28 rs12979860 and rs8099917
2.4. Statistical Analysis
3. Results
4. Discussion
5. Study Limitations
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Park, J.-W.; Chen, M.; Colombo, M.; Roberts, L.R.; Schwartz, M.; Schwartz, M.; Chen, P.J.; Kudo, M.; Johnson, P.; Wagner, S.; et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015, 35, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL-EORTC Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, R.H.; Dusheiko, G. Natural history of hepatitis C. J. Hepatol. 2014, 61 (Suppl. 1), S58–S68. [Google Scholar] [CrossRef] [PubMed]
- Goossens, N.; Hoshida, Y. Hepatitis C virus-induced hepatocellular carcinoma. Clin. Mol. Hepatol. 2015, 21, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Fabris, C.; Falleti, E.; Cussigh, A.; Bitetto, D.; Fontanini, E.; Bignulin, S.; Cmet, S.; Fornasiere, E.; Fumolo, E.; Fangazio, S.; et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: Role in the course of chronic viral hepatitis and the development of HCC. J. Hepatol. 2011, 54, 716–722. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B.; Kramer, J.; Duan, Z.; Kanwal, F. Epidemiology and outcomes of hepatitis C infection in elderly US Veterans. J. Viral Hepat. 2016, 23, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Khattab, M.A.; Abdelghany, H.M.; Ramzy, M.M.; Khairy, R.M. Impact of IL28B gene polymorphisms rs8099917 and rs12980275 on response to pegylated interferon-α/ribavirin therapy in chronic hepatitis C genotype 4 patients. J. Biomed. Res. 2016, 30, 40–45. [Google Scholar] [CrossRef]
- McGlynn, K.A.; London, W.T. The global epidemiology of hepatocellular carcinoma: Present and future. Clin. Liver Dis. 2011, 15, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, S.; Park, S.H. The epidemiology of hepatocellular cancer: From the perspectives of public health problem to tumor biology. J. Gastroenterol. 2009, 44 (Suppl. 19), 96–101. [Google Scholar] [CrossRef] [PubMed]
- Liakina, V.; Valantinas, J. Anti-HCV prevalence in the general population of Lithuania. Med. Sci. Monit. 2012, 18, PH28–PH35. [Google Scholar] [CrossRef] [PubMed]
- Vallet-Pichard, A.; Mallet, V.; Nalpas, B.; Verkarre, V.; Nalpas, A.; Dhalluin-Venier, V.; Fontaine, H.; Pol, S. FIB-4: An inexpensive and accurate marker of fibrosis in HCV infection. Comparison with liver biopsy and fibrotest. Hepatology 2007, 46, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Kim, S.A.; Park, Y.N.; Cheong, J.Y.; Kim, H.S.; Park, J.Y.; Cho, S.W.; Han, K.H.; Chon, C.Y.; Moon, Y.M.; et al. Noninvasive models to predict liver cirrhosis in patients with chronic hepatitis B. Liver Int. 2007, 27, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaoki, Y.; Aikata, H.; Nakano, N.; Shinohara, F.; Nakamura, Y.; Hatooka, M.; Morio, K.; Kan, H.; Fujino, H.; Kobayashi, T.; et al. Development of hepatocellular carcinoma in patients with hepatitis C virus infection who achieved sustained virological response following interferon therapy: A large-scale, long-term cohort study. J. Gastroenterol. Hepatol. 2016, 31, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Koike, K. Molecular Basis of Hepatitis C Virus–Associated Hepatocarcinogenesis: Lessons from Animal Model Studies. Clin. Gastroenterol. Hepatol. 2005, 3 (Suppl. 2), S132–S135. [Google Scholar] [CrossRef]
- Mitchell, J.K.; Lemon, S.M.; McGivern, D.R. How do persistent infections with hepatitis C virus cause liver cancer? Curr. Opin. Virol. 2015, 14, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singal, A.G.; Pillai, A.; Tiro, J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: A meta-analysis. PLoS Med. 2014, 11, e1001624. [Google Scholar] [CrossRef] [PubMed]
- Thein, H.H.; Yi, Q.; Dore, G.J.; Krahn, M.D. Estimation of stage-specific fibrosis progression rates in chronic hepatitis C virus infection: A meta-analysis and meta-regression. Hepatology 2008, 48, 418–431. [Google Scholar] [CrossRef] [PubMed]
- Tabrizian, P.; Jibara, G.; Shrager, B.; Schwartz, M.; Roayaie, S. Recurrence of hepatocellular cancer after resection: Patterns, treatments, and prognosis. Ann. Surg. 2015, 261, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Kashuba, E.; Kashuba, V.; Sandalova, T.; Klein, G.; Szekely, L. Epstein-Barr virus encoded nuclear protein EBNA-3 binds a novel human uridine kinase/uracil phosphoribosyltransferase. BMC Cell Biol. 2002, 3, 23. [Google Scholar] [CrossRef]
- Ahmed, N. Enzymes of de novo and salvage pathways for pyrimidine biosynthesis in normal colon, colon carcinoma, and xenografts. Cancer 1984, 54, 1370–1373. [Google Scholar] [CrossRef]
- Shen, F.; Look, K.; Yeh, Y.; Weber, G. Increased uridine kinase (ATP: Uridine 5′-phosphotransferase; EC 2.7.1.48) activity in human and rat tumors. Cancer Biochem. Biophys. 1998, 16, 1–15. [Google Scholar] [PubMed]
- Yuh, I.; Yaoi, T.; Watanase, S.; Okajima, S.; Hirasawa, Y.; Fushiki, S. Up-regulated uridine kinase gene identified by RLCS in the ventral horn after crush injury to rat sciatic nerves. Biochem. Biophys. Res. Commun. 1999, 266, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, E.C.; Kornbluth, J. Downregulation of uridine-cytidine kinase like-1 decreases proliferation and enhances tumor susceptibility to lysis by apoptotic agents and natural killer cells. Apoptosis 2009, 14, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Asahina, Y.; Tsuchiya, K.; Tamaki, N.; Hirayama, I.; Tanaka, T.; Sato, M.; Yasui, Y.; Hosokawa, T.; Ueda, K.; Kuzuya, T.; et al. Effect of aging on risk for hepatocellular carcinoma in chronic hepatitis C virus infection. Hepatology 2010, 52, 518–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prokunina-Olsson, L.; Muchmore, B.; Tang, W.; Pfeiffer, R.M.; Park, H.; Dickensheets, H.; Yasui, Y.; Hosokawa, T.; Ueda, K.; Kuzuya, T.; et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013, 45, 164–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robek, M.D.; Boyd, B.S.; Chisari, F.V. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 2005, 79, 3851–3854. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.L. Preclinical and clinical development of pegylated interferon-lambda 1 in chronic hepatitis C. J. Interferon Cytokine Res. 2010, 30, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.J.; Shiffman, M.L.; Zaman, A.; Yoffe, B.; de la Torre, A.; Flamm, S.; Gordon, S.C.; Marotta, P.; Vierling, J.M.; Carlos Lopez-Talavera, J.; et al. Phase 1b study of pegylated interferon-lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology 2010, 52, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Asahina, Y.; Tsuchiya, K.; Nishimura, T.; Muraoka, M.; Suzuki, Y.; Tamaki, N.; Yasui, Y.; Hosokawa, T.; Ueda, K.; Nakanishi, H.; et al. Genetic variation near interleukin 28B and the risk of hepatocellular carcinoma in patients with chronic hepatitis C. J. Gastroenterol. 2014, 49, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Kato, N.; Tateishi, R.; Muroyama, R.; Kowatari, N.; Li, W.; Goto, K.; Otsuka, M.; Shiina, S.; Yoshida, H.; et al. IL28B minor allele is associated with a younger age of onset of hepatocellular carcinoma in patients with chronic hepatitis C virus infection. J. Gastroenterol. 2014, 49, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Hodo, Y.; Honda, M.; Tanaka, A.; Nomura, Y.; Arai, K.; Yamashita, T.; Sakai, Y.; Yamashita, T.; Mizukoshi, E.; Sakai, A.; et al. Association of interleukin-28B genotype and hepatocellular carcinoma recurrence in patients with chronic hepatitis C. Clin. Cancer Res. 2013, 19, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Joshita, S.; Umemura, T.; Katsuyama, Y.; Ichikawa, Y.; Kimura, T.; Morita, S.; Kamijo, A.; Komatsu, M.; Ichijo, T.; Matsumoto, A.; et al. Association of IL28B gene polymorphism with development of hepatocellular carcinoma in Japanese patients with chronic hepatitis C virus infection. Hum. Immunol. 2012, 73, 298–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indolfi, G.; Azzari, C.; Resti, M. Polymorphisms in the IFNL3/IL28B gene and hepatitis C: From adults to children. World J. Gastroenterol. 2014, 20, 9245–9252. [Google Scholar] [CrossRef] [PubMed]
- Langhans, B.; Kupfer, B.; Braunschweiger, I.; Arndt, S.; Schulte, W.; Nischalke, H.D.; Nattermann, J.; Oldenburg, J.; Sauerbruch, T.; Spengler, U. Interferon-lambda serum levels in hepatitis, C. J. Hepatol. 2011, 54, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; McLeod, D.; Kelaeng, K.S.; Mangia, A.; Berg, T.; Thabet, K.; Irving, W.L.; Dore, G.J.; Sheridan, D.; Grønbæk, H.; et al. IFN-λ3, not IFN-λ4, likely mediates IFNL3-IFNL4 haplotype-dependent hepatic inflammation and fibrosis. Nat. Genet. 2017, 49, 795–800. [Google Scholar] [CrossRef] [PubMed]
- López-Rodríguez, R.; Hernández-Bartolomé, Á.; Borque, M.J.; Rodríguez-Muñoz, Y.; Martín-Vílchez, S.; García-Buey, L.; González-Moreno, L.; Real-Martínez, Y.; de Rueda, P.M.; Salmerón, J.; et al. Interferon-related genetic markers of necroinflammatory activity in chronic hepatitis C. PLoS ONE 2017, 12, e0180927. [Google Scholar] [CrossRef] [PubMed]
- Steen, H.C.; Gamero, A.M. Interferon-lambda as a potential therapeutic agent in cancer treatment. J. Interferon Cytokine Res. 2010, 30, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; George, J. Targeting IFN-λ: Therapeutic implications. Expert Opin. Ther. Targets 2016, 20, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Jiang, C.; Peng, Z.; Liu, B.; Hu, W.; Wang, Y.; Lin, M.; Lu, M.; Kuang, M. Local recurrence after radiofrequency ablation of hepatocellular carcinoma: Treatment choice and outcome. J. Gastrointest. Surg. 2015, 19, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Humphreys, I.; Flaxman, A.; Brown, A.; Cooke, G.S.; Pybus, O.G.; Barnes, E. Global Distribution and Prevalence of Hepatitis C Virus Genotypes. Hepatology 2015, 61, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Gower, E.; Estes, C.; Blach, S.; Razavi-Shearer, K.; Razavi, H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol. 2014, 61 (Suppl. 1), S45–S57. [Google Scholar] [CrossRef] [PubMed]
- Al-Qahtani, A.; Al-Anazi, M.; Abdo, A.A.; Sanai, F.M.; Al-Hamoudi, W.; Alswat, K.A.; Al-Ashgar, H.I.; Khan, M.Q.; Albenmousa, A.; Khalaf, N.; et al. Correlation between genetic variations and serum level of interleukin 28B with virus genotypes and disease progression in chronic hepatitis C virus infection. J. Immunol. Res. 2015, 2015, 768470. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yu, G.; Li, Z.; Liang, H. Influence of interleukin-28B polymorphism on progression to hepatitis virus-induced hepatocellular carcinoma. Tumour Biol. 2014, 35, 8757–8763. [Google Scholar] [CrossRef] [PubMed]
- Suo, G.J.; Zhao, Z.X. Association of the interleukin-28B gene polymorphism with development of hepatitis virus-related hepatocellular carcinoma and liver cirrhosis: A meta-analysis. Genet. Mol. Res. 2013, 12, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- King, L.Y.; Johnson, K.B.; Zheng, H.; Wei, L.; Gudewicz, T.; Hoshida, Y.; Corey, K.E.; Ajayi, T.; Ufere, N.; Baumert, T.F.; et al. Host genetics predict clinical deterioration in HCV-related cirrhosis. PLoS ONE 2014, 9, e114747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eurich, D.; Boas-Knoop, S.; Bahra, M.; Neuhaus, R.; Somasundaram, R.; Neuhaus, P.; Neumann, U.; Seehofer, D. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation 2012, 93, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Kashuba, E.; Bailey, J.; Allsup, D.; Cawkwell, L. The kinin-kallikrein system: Physiological roles, pathophysiology and its relationship to cancer biomarkers. Biomarkers 2013, 18, 279–296. [Google Scholar] [CrossRef] [PubMed]
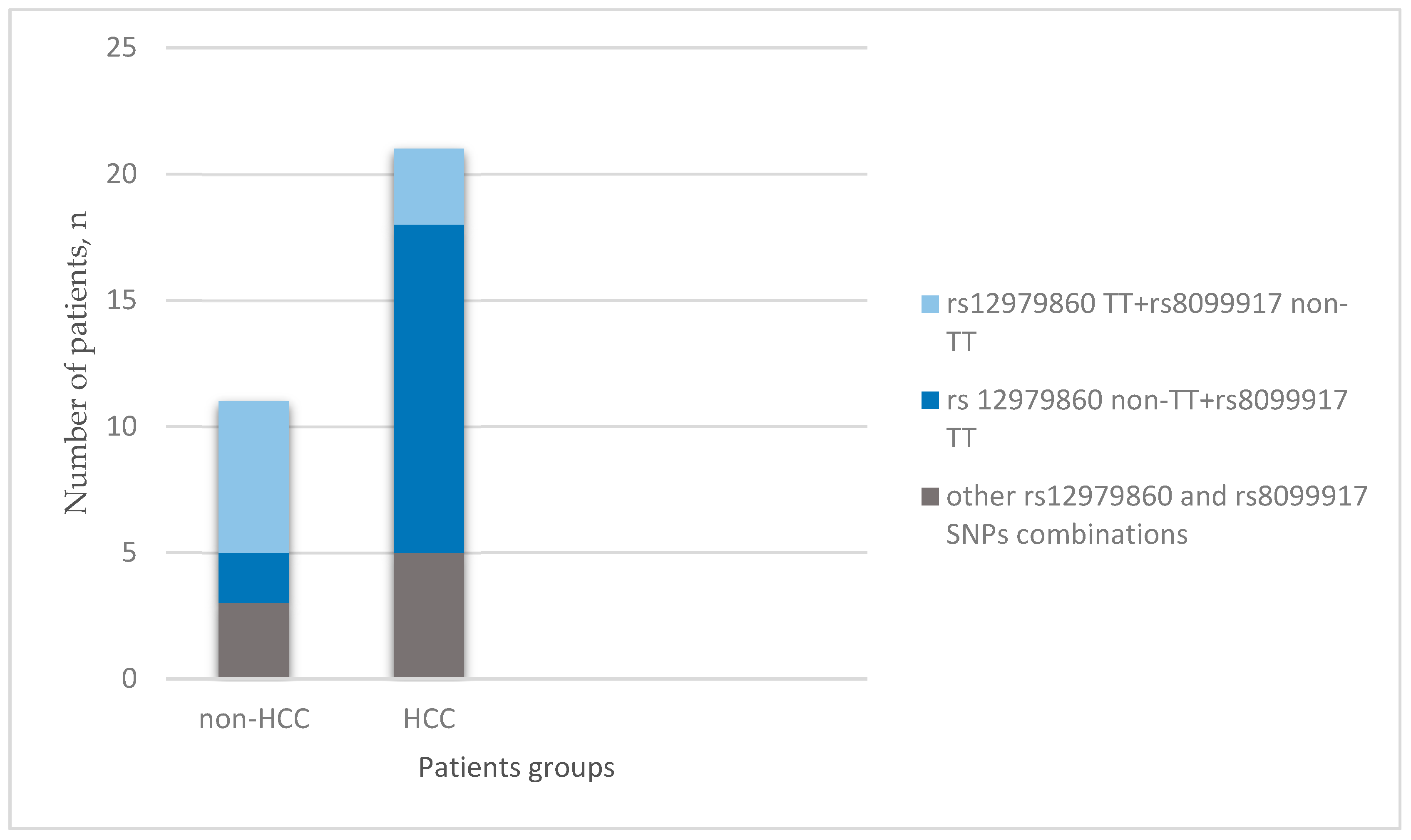
| Characteristics | HCC (n = 21) | Non-HCC (n = 11) | p Value |
|---|---|---|---|
| Age, mean ± SD, years | 55.14 ± 5.46 | 48.36 ± 8.96 | 0.012 |
| Gender, n (%): | |||
| Women | 4 (19) | 4 (36.4) | 0.434 |
| Men | 17 (81) | 7 (63.6) | |
| BMI, mean ± SD, kg/m2 | 25.75 ± 4.29 | 26.36 ± 4.15 | 0.706 |
| HCV genotype, n (%): | |||
| G1 | 12 (57.1) | 9 (81.8) | 0.307 |
| G2 | 2 (9.5) | 0 (0) | |
| G3 | 7 (33.3) | 2 (18.2) | |
| Hepatitis C treatment, n (%) | 14 (66.7) | 9 (81.8) | 0.506 |
| HAI, mean ± SD, grade | 6.52 ±1.86 | 6.64 ±2.34 | 0.883 |
| Steatosis, mean ± SD, % | 13.8 ± 17.1 | 7.7 ± 9.8 | 0.287 |
| UCKL-1, mean ± SD, relative units | 105 ± 58.52 | 43.18 ± 31.01 | 0.003 |
| IL28B Polymorphisms | HCC (n = 21) | Non-HCC (n = 11) | p Value |
|---|---|---|---|
| rs12979860, n (%): | |||
| CC | 9 (42.9) | 1 (9.1) | 0.481 |
| TC | 9 (42.9) | 4 (36.4) | |
| TT | 3 (14.3) | 6 (54.5) | |
| rs8099917, n (%): | |||
| TT | 13 (61.9) | 2 (18.2) | 0.015 |
| TG | 5 (23.8) | 7 (63.6) | |
| GG | 3 (14.3) | 2 (18.2) |
| Variable | OR | 95% Confidence Interval | p |
|---|---|---|---|
| UCKL-1 | 1.0279 | 1.0066–1.0496 | 0.0100 |
| IL28B rs8099917 TT | 7.3125 | 1.2490–42.8134 | 0.0273 |
| IL28B rs8099917 TG | 0.1786 | 0.0365–0.8728 | 0.0333 |
| IL28B rs12979860 TT | 0.1389 | 0.0253–0.7631 | 0.0231 |
| IL28B rs12979860 CC | 7.5000 | 0.8067–69.7326 | 0.0765 |
| Age | 1.1882 | 1.0144–1.3919 | 0.0326 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buivydiene, A.; Liakina, V.; Kashuba, E.; Norkuniene, J.; Jokubauskiene, S.; Gineikiene, E.; Valantinas, J. Impact of the Uridine–Cytidine Kinase Like-1 Protein and IL28B rs12979860 and rs8099917 SNPs on the Development of Hepatocellular Carcinoma in Cirrhotic Chronic Hepatitis C Patients—A Pilot Study. Medicina 2018, 54, 67. https://0-doi-org.brum.beds.ac.uk/10.3390/medicina54050067
Buivydiene A, Liakina V, Kashuba E, Norkuniene J, Jokubauskiene S, Gineikiene E, Valantinas J. Impact of the Uridine–Cytidine Kinase Like-1 Protein and IL28B rs12979860 and rs8099917 SNPs on the Development of Hepatocellular Carcinoma in Cirrhotic Chronic Hepatitis C Patients—A Pilot Study. Medicina. 2018; 54(5):67. https://0-doi-org.brum.beds.ac.uk/10.3390/medicina54050067
Chicago/Turabian StyleBuivydiene, Arida, Valentina Liakina, Elena Kashuba, Jolita Norkuniene, Skirmante Jokubauskiene, Egle Gineikiene, and Jonas Valantinas. 2018. "Impact of the Uridine–Cytidine Kinase Like-1 Protein and IL28B rs12979860 and rs8099917 SNPs on the Development of Hepatocellular Carcinoma in Cirrhotic Chronic Hepatitis C Patients—A Pilot Study" Medicina 54, no. 5: 67. https://0-doi-org.brum.beds.ac.uk/10.3390/medicina54050067





