Antimicrobial and Antioxidant Polyketides from a Deep-Sea-Derived Fungus Aspergillus versicolor SH0105
Abstract
:1. Introduction
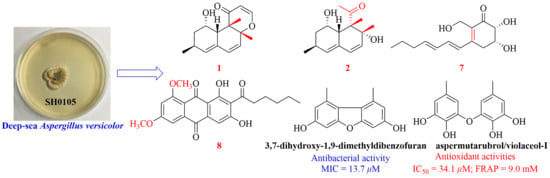
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction, and Isolation
3.4. ECD Calculations
3.5. Biological Assays
3.5.1. Antimicrobial Assay
3.5.2. Antioxidant Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; He, X.Q.; Wu, G.W.; Liu, C.C.; Lu, C.J.; Gu, Q.Q.; Che, Q.; Zhu, T.J.; Zhang, G.J.; Li, D.H. Aniline-tetramic acids from the deep-sea-derived fungus Cladosporium sphaerospermum L3P3 cultured with the HDAC inhibitor SAHA. J. Nat. Prod. 2018, 81, 1651–1657. [Google Scholar] [CrossRef]
- Wu, J.S.; Yao, G.S.; Shi, X.H.; Rehman, S.U.; Xu, Y.; Fu, X.M.; Zhang, X.L.; Liu, Y.; Wang, C.Y. Epigenetic agents trigger the production of bioactive nucleoside derivatives and bisabolane sesquiterpenes from the marine-derived fungus Aspergillus versicolor. Front. Microbiol. 2020, 11, 85. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.L.; Li, Y.X.; Banakar, S.P.; Liu, L.; Shao, C.L.; Li, Z.Y.; Wang, C.Y. New metabolites from the co-culture of marine-derived actinomycete Streptomyces rochei MB037 and fungus Rhinocladiella similis 35. Front. Microbiol. 2019, 10, 915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.Y.; Wu, J.T.; Shao, C.L.; Li, Z.Y.; Chen, M.; Wang, C.Y. Co-culture: Stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms. Mar. Life. Sci. Technol. 2020. [Google Scholar] [CrossRef]
- Li, Z.Y.; Zhu, D.Y.; Shen, Y.M. Discovery of novel bioactive natural products driven by genome mining. Drug. Discov. Ther. 2018, 12, 318–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Liu, Z.Y.; Sun, C.L.; Shao, M.W.; Ma, J.Y.; Wei, X.Y.; Zhang, T.Y.; Li, W.J.; Ju, J.H. Discovery and biosynthesis of atrovimycin, an antitubercular and antifungal cyclodepsipeptide featuring vicinal-dihydroxylated cinnamic acyl chain. Org. Lett. 2019, 21, 2634–2638. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Y.; Liu, H.S.; Zhu, W.M. New natural products from the marine-derived Aspergillus fungi—A review. Acta Microbiol. Sin. 2016, 56, 331–362. [Google Scholar] [CrossRef]
- Wang, K.W.; Ding, P. New bioactive metabolites from the marine-derived fungi Aspergillus. Mini-Rev. Med. Chem. 2018, 18, 1072–1094. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.; Lloyd, K.; Miller, B.R.; Palladino, M.A.; Kiso, Y.; Hayashi, Y.; Neuteboom, S.T.C. NPI-2358 is a tubulin-depolymerizing agent: In-vitro evidence for activity as a tumor vascular-disrupting agent. Anti-Cancer Drug. 2006, 17, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ingavat, N.; Dobereiner, J.; Wiyakrutta, S.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Aspergillusol A, an α-glucosidase inhibitor from the marine-derived fungus Aspergillus aculeatus. J. Nat. Prod. 2009, 72, 2049–2052. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Wei, X.Y.; Qin, X.C.; Tian, X.P.; Li, K.M.; Zhou, X.F.; Yang, X.W.; Wang, F.Z.; Zhang, T.Y.; Tu, Z.C.; et al. Antiviral merosesquiterpenoids produced by the Antarctic fungus Aspergillus ochraceopetaliformis SCSIO 05702. J. Nat. Prod. 2016, 79, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Cimino, P.J.; Huang, L.; Du, L.H.; Wu, Y.P.; Bishop, J.; Dalsing-Hernandez, J.; Kotlarczyk, K.; Gonzales, P.; Carew, J.; Nawrocki, S.; et al. Plinabulin, an inhibitor of tubulin polymerization, targets KRAS signaling through disruption of endosomal recycling. Biomed. Rep. 2019, 10, 218–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.Y.; Zhang, Y.; Xu, X.Y.; Qi, S.H. Diverse deep-sea fungi from the south China sea and their antimicrobial activity. Curr. Microbiol. 2013, 67, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Xue, Y.R.; Liu, C.H. A brief review of bioactive metabolites derived from deep-sea fungi. Mar. Drugs. 2015, 13, 4594–4616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, Z.E.; Brimble, M.A. Molecules derived from the extremes of life: A decade later. Nat. Prod. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; He, W.J.; Huang, X.L.; Tian, X.P.; Liao, S.R.; Yang, B.; Wang, F.Z.; Zhou, X.J.; Liu, Y.H. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agr. Food. Chem. 2016, 64, 2910–2916. [Google Scholar] [CrossRef]
- Wang, J.; Yao, Q.F.; Amin, M.; Nong, X.H.; Zhang, X.Y.; Qi, S.H. Penicillenols from a deep-sea fungus Aspergillus restrictus inhibit Candida albicans biofilm formation and hyphal growth. J. Antibiot. 2017, 70, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.Y.; Li, M.J.; Lin, Y.Z.; Du, S.W.; Liu, Z.Y.; Ju, J.H.; Suzuki, H.; Sawada, M.; Umezawa, K. Inhibition of cellular inflammatory mediator production and amelioration of learning deficit in flies by deep sea Aspergillus derived cyclopenin. J. Antibiot. 2020, 73, 622–629. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, Y.Y.; Shao, C.L.; Wang, C.Y. Metabolites from marine invertebrates and their symbiotic microorganisms: Molecular diversity discovery, mining, and application. Mar. Life. Sci. Technol. 2019, 1, 60–94. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Xu, Y.; Shao, C.L.; Yang, R.Y.; Zheng, C.J.; Chen, Y.Y.; Fu, X.M.; Qian, P.Y.; She, Z.G.; Voogd, N.J.D.; et al. Antibacterial bisabolane-type sesquiterpenoids from the sponge-derived fungus Aspergillus sp. Mar. Drugs. 2012, 10, 234–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Shao, C.L.; Meng, H.; She, Z.G.; Wang, C.Y. Anti-respiratory syncytial virus prenylated dihydroquinolone derivatives from the gorgonian-derived fungus Aspergillus sp. XS-20090B15. J. Nat. Prod. 2014, 77, 2720–2724. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.M.; Liang, T.M.; Guo, Z.Y.; Wang, C.Y.; Shao, C.L. Discovery, absolute assignments, and total synthesis of asperversiamides A–C and their potent activity against Mycobacterium marinum. Chem. Commun. 2019, 55, 1104–1107. [Google Scholar] [CrossRef] [PubMed]
- An, C.L.; Kong, F.D.; Ma, Q.Y.; Xie, Q.Y.; Yuan, J.Z.; Zhou, L.M.; Dai, H.F.; Yu, Z.F.; Zhao, Y.X. Chemical constituents of the marine-derived fungus Aspergillus sp. SCS-KFD66. Mar. Drugs. 2018, 16, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuyama, K.; Tsukihara, T.; Katsube, Y. Structure of versiol, a new metabolite from Aspergillus versicolor. Tetrahedron Lett. 1976, 3, 189–190. [Google Scholar] [CrossRef]
- Cho, N.; Ransom, T.T.; Sigmund, J.; Cichewicz, R.H.; Goetz, M.; Beutler, J.A. Growth inhibition of colon cancer and melanoma cells by versiol derivatives from a Paraconiothyrium species. J. Nat. Prod. 2017, 80, 2037–2044. [Google Scholar] [CrossRef]
- Fujii, Y.; Asahara, M.; Ichinoe, M.; Nakajima, H. Fungal melanin inhibitor and related compounds from Penicillium decumbens. Phytochemistry 2002, 60, 703–708. [Google Scholar] [CrossRef]
- Ola, A.R.B.; Tawo, B.D.; Belli, H.L.L.; Proksch, P.; Tommy, D.; Hakim, E.H. A new antibacterial polyketide from the endophytic fungi Aspergillus fumigatiaffinis. Nat. Prod. Commun. 2018, 13, 1573–1574. [Google Scholar] [CrossRef]
- Yamazaki, M.; Satoh, Y.; Maebayashi, Y.; Horie, Y. Monoamine oxidase inhibitors from a fungus, Emericella navahoensis. Chem. Pharm. Bull. 1988, 36, 670–675. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.L.; Wang, C.Y.; Wei, M.Y.; Li, S.D.; She, Z.G.; Gu, Y.C.; Lin, Y.C. Structural and spectral assignments of six anthraquinone derivatives from the mangrove fungus (ZSUH-36). Magn. Reson. Chem. 2008, 46, 886–889. [Google Scholar] [CrossRef]
- Steyn, P.S.; Vleggaar, R.; Wessels, P.L. Structure and carbon-13 nuclear magnetic resonance assignments of versiconal acetate, versiconol acetate, and versiconol, metabolites from cultures of Aspergillus parasiticus treated with dichlorvos. J. Chem. Soc. Perkin Trans. 1979, 1, 451–459. [Google Scholar] [CrossRef]
- Lee, Y.M.; Li, H.Y.; Hong, J.; Cho, H.Y.; Bae, K.S.; Kim, M.A.; Kim, D.K.; Jung, J.H. Bioactive metabolites from the sponge-derived fungus Aspergillus versicolor. Arch. Pharm. Res. 2010, 33, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.H.; Cole, R.J. Carbon-13 nuclear magnetic resonance studies of fungal metabolites, aflatoxins, and sterigmatocystins. J. Org. Chem. 1977, 42, 112–114. [Google Scholar] [CrossRef] [PubMed]
- Tanahashia, T.; Takenaka, Y.; Nagakura, N.; Hamada, N. Dibenzofurans from the cultured lichen mycobionts of Ecanora cinereocarnea. Phytochemistry 2001, 8, 1129–1134. [Google Scholar] [CrossRef]
- Taniguchi, M.; Kaneda, N.; Shibata, K.; Kamikawa, T. Isolation and biological activity of aspermutarubrol, a self-growth inhibitor from Aspergillus sydowi. Agr. Biol. Chem. 1978, 42, 1629–1630. [Google Scholar] [CrossRef] [Green Version]
- Nakamaru, T.; Shiojiri, H.; Kawai, K.; Nozawa, Y.; Maebayashi, Y.; Yamazaki, M. The effects of toxic metabolites violaceol I and II from Emericella violacea on mitochondrial respiration. Mycotoxins. 1984, 19, 30–33. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, M.; Chakraborty, A.; Mukherjee, P. Antibiotic resistance: The global crisis. J. Pharm. Res 2020, 9, 12–16. [Google Scholar] [CrossRef]
- Dugassa, J.; Shukuri, N. Review on antibiotic resistance and its mechanism of development. J. Health Med. Nurs. 2017, 1, 1–17. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Bruhn, T.; Schaumloffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Aktumsek, A.; Zengin, G.; Guler, G.O.; Cakmak, Y.S.; Duran, A. Antioxidant potentials and anticholinesterase activities of methanolic andaqueous extracts of three endemic Centaurea L. species. Food. Chem. Toxicol. 2013, 55, 290–296. [Google Scholar] [CrossRef]






| No. | 1 a | 2 b | 7 b | |||
|---|---|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, Type | δH (J in Hz) | |
| 1 | 67.5, CH | 5.20, m | 67.3, CH | 4.22, m | 199.6, C | |
| 2 | 38.8, CH2 | 2.00, m; 1.30, m | 40.8, CH2 | 1.85, m; 1.22, m | 76.8, CH | 4.31, d (3.0) |
| 3 | 25.9, CH | 2.56, m | 26.9, CH | 2.53, m | 70.9, CH | 4.41, m |
| 4 | 136.6, CH | 5.71, brs | 134.9, CH | 5.58, brs | 33.8, CH2 | 3.01, dd (18.2, 3.5)2.90, dd (18.2, 3.5) |
| 5 | 130.3, C | 132.7, C | 151.0, C | |||
| 6 | 130.5, CH | 6.00, d (9.8) | 129.6, CH | 5.94, d (9.8) | 132.5, C | |
| 7 | 127.6, CH | 5.64, d (9.8) | 134.1, CH | 5.35, d (9.8) | 54.5, CH2 | 4.40, d (11.6)4.58, d (11.6) |
| 8 | 86.2, C | 75.2, C | 128.4, CH | 6.88, m | ||
| 9 | 49.4, C | 58.9, C | 139.3, CH | 6.87, m | ||
| 10 | 41.9, CH | 2.65, q (3.3) | 43.3, CH | 2.88, q (3.3) | 132.3, CH | 6.33, ddq (15.1, 8.4, 1.4) |
| 11 | 200.2, C | 216.3, C | 142.3, CH | 6.11, dt (15.1, 7.2) | ||
| 12 | 106.5, CH | 5.31, d (5.9) | 30.8, CH3 | 2.28, s | 36.1, CH2 | 2.19, dq (7.2, 1.4) |
| 13 | 158.4, CH | 7.05, d (5.9) | 26.5, CH3 | 1.12, s | 23.3, CH2 | 1.50, qt (7.4, 1.4) |
| 14 | 21.0, CH3 | 1.47, s | 15.2, CH3 | 1.44, s | 14.0, CH3 | 0.96, t (7.4) |
| 15 | 17.2, CH3 | 1.28, s | 21.6, CH3 | 1.02, d (7.1) | ||
| 16 | 21.2, CH3 | 1.04, d (7.1) | ||||
| No. | δC, type | δH (J in Hz) | No. | δC, type | δH (J in Hz) |
|---|---|---|---|---|---|
| 1 | 163.1, C | 13 | 183.5, C | ||
| 2 | 104.6, CH | 6.99, d (2.5) | 14 | 113.8, C | |
| 3 | 164.9, C | 15 | 203.2, C | ||
| 4 | 104.9, CH | 7.24, d (2.5) | 16 | 43.7, CH2 | 2.74, t (7.3) |
| 5 | 133.7, C | 17 | 22.6, CH2 | 1.54, dd (8.5, 5.6) | |
| 6 | 181.8, C | 18 | 30.7, CH2 | 1.25, m | |
| 7 | 136.4, C | 19 | 21.9, CH2 | 1.25, m | |
| 8 | 106.6, CH | 7.08, s | 20 | 13.8, CH3 | 0.83, t (7.0) |
| 9 | 163.0, C | 1-OCH3 | 56.6, CH3 | 3.91, s | |
| 10 | 121.6, C | 3-OCH3 | 56.2, CH3 | 3.99, s | |
| 11 | 161.3, C | 11-OH | 13.9, s | ||
| 12 | 109.1, C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.-J.; Peng, X.-Y.; Zhang, Y.-H.; Liu, Z.-Q.; Li, X.; Gu, Y.-C.; Shao, C.-L.; Han, Z.; Wang, C.-Y. Antimicrobial and Antioxidant Polyketides from a Deep-Sea-Derived Fungus Aspergillus versicolor SH0105. Mar. Drugs 2020, 18, 636. https://0-doi-org.brum.beds.ac.uk/10.3390/md18120636
Yang L-J, Peng X-Y, Zhang Y-H, Liu Z-Q, Li X, Gu Y-C, Shao C-L, Han Z, Wang C-Y. Antimicrobial and Antioxidant Polyketides from a Deep-Sea-Derived Fungus Aspergillus versicolor SH0105. Marine Drugs. 2020; 18(12):636. https://0-doi-org.brum.beds.ac.uk/10.3390/md18120636
Chicago/Turabian StyleYang, Lu-Jia, Xiao-Yue Peng, Ya-Hui Zhang, Zhi-Qing Liu, Xin Li, Yu-Cheng Gu, Chang-Lun Shao, Zhuang Han, and Chang-Yun Wang. 2020. "Antimicrobial and Antioxidant Polyketides from a Deep-Sea-Derived Fungus Aspergillus versicolor SH0105" Marine Drugs 18, no. 12: 636. https://0-doi-org.brum.beds.ac.uk/10.3390/md18120636