Recombinant Expression of Thrombolytic Agent Reteplase in Marine Microalga Tetraselmis subcordiformis (Chlorodendrales, Chlorophyta)
Abstract
:1. Introduction
2. Results
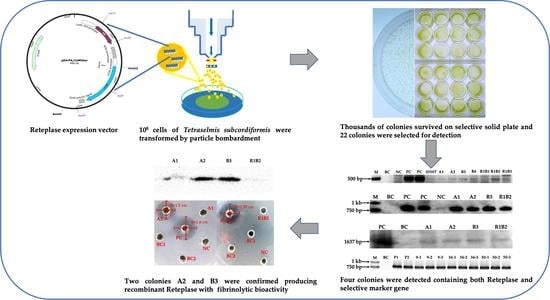
2.1. Biolistic Transformation and Basta Selection
2.2. Detection of the bar and rt-PA Gene Integration
2.3. Purification of Recombinant Reteplase, Western Blotting and the Fibrinolysis Activity Assay
3. Discussion
4. Materials and Methods
4.1. Algae Culture
4.2. Plasmid Vector for Transformation
4.3. Biolistic Transformation
4.4. Basta Selection for Positive Transformants
4.5. PCR detection and Southern Blotting
4.6. Purification of Recombinant Reteplase and Western Blotting
4.7. Fibrinolysis Activity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Qiu, Z.; Zhan, D.; Jian, Y.; Ye, H. Study on the applying of dried Platymonas sp. in the seed rearing of the bay scallop. Shandong Fish. 2003, 20, 37–39. [Google Scholar]
- Zheng, Y.; Wang, G.; Li, S. Impacts of Phaeodactylum tricornutum and Platymonas subcordiformis on fecundity, survival and fatty acid of Apocyclops borneoensis. J. Xiamen Univ. 2012, 51, 402–409. [Google Scholar]
- Guan, Y.; Deng, M.; Yu, X.; Zhang, W. Two-stage photo-biological production of hydrogen by marine green alga Platymonas subcordiformis. Biochem. Eng. J. 2004, 19, 69–73. [Google Scholar] [CrossRef]
- Yao, C.; Ai, J.; Cao, X.; Xue, S.; Zhang, W. Enhancing starch production of a marine green microalga Tetraselmis subcordiformis through nutrient limitation. Bioresour. Technol. 2012, 118, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, R.; Zhuang, H.; Ou, L.; Huang, X. FT Raman spectra study of Platymonas subcordiformis. Spectrosc. Spectr. Anal. 2007, 27, 81–83. [Google Scholar]
- Xu, D.; Gao, Z.; Li, F.; Fan, X.; Zhang, X.; Ye, N.; Mou, S.; Liang, C.; Li, D. Detection and quantitation of lipid in the microalga Tetraselmis subcordiformis (Wille) Butcher with BODIPY 505/515 staining. Bioresour. Technol. 2013, 127, 386–390. [Google Scholar] [CrossRef]
- Ma, R.; Wang, B.; Chua, E.T.; Zhao, X.; Lu, K.; Ho, S.; Shi, X.; Liu, L.; Xie, Y.; Lu, Y.; et al. Comprehensive utilization of marine microalgae for enhanced co-production of multiple compounds. Mar. Drugs 2020, 18, 467. [Google Scholar] [CrossRef]
- Borowitzka, M.A. Microalgae for aquaculture: Opportunities and constraints. J. Appl. Phycol. 1997, 9, 393–401. [Google Scholar] [CrossRef]
- Muller-Feuga, A. The role of microalgae in aquaculture: Situation and trends. J. Appl. Phycol. 2000, 12, 527–534. [Google Scholar] [CrossRef]
- Cui, Y.; Jiang, P.; Wang, J.; Li, F.; Chen, Y.; Zheng, G.; Qin, S. Genetic transformation of Platymonas (Tetraselmis) subcordiformis (Prasinophyceae, Chlorophyta) using particle bombardment and glass-bead agitation. Chin. J. Oceanol. Limnol. 2012, 30, 471–475. [Google Scholar] [CrossRef]
- Cui, Y.; Qin, S.; Jiang, P. Chloroplast transformation of Platymonas (Tetraselmis) subcordiformis with the bar gene as selectable marker. PLoS ONE 2014, 9, e98607. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Qu, L.; Zhao, J.; Qin, S. Transient expression of the enhanced green fluorescence protein (egfp) gene in Tetraselmis subcordiformis (Chlorodendrales, Chlorophyta) with three exogenous promoters. Phycologia 2016, 55, 564–567. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Wang, J.; Jiang, P.; Bian, S.; Qin, S. Transformation of Platymonas (Tetraselmis) subcordiformis (Prasinophyceae, Chlorophyta) by agitation with glass beads. World J. Microb. Biot. 2010, 26, 1653–1657. [Google Scholar] [CrossRef]
- Pennica, D.; Holmes, W.E.; Kohr, W.J.; Harkins, R.N.; Vehar, G.A.; Ward, C.A.; Bennett, W.F.; Yelverton, E.; Seeburg, P.H.; Heyneker, H.L.; et al. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature 1983, 20, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Kohnert, U.; Rudolph, R.; Verheijen, J.H.; Jacoline, E.; Weening-Verhoeff, D.; Anne, S.; Ulrich, O.; Ulrich, M.; Helmut, L.; Heinrich, P. Biochemical properties of the kringle 2 and protease domains are maintained in the refolded t-PA deletion variant BM 06.022. Protein. Eng. Des. Sel. 1992, 5, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Stürzebecher, J.; Neumann, U.; Kohnert, U.; Kresse, G.B.; Fischer, S. Mapping of the catalytic site of CHO-t-PA and the t-PA variant BM 06.022 by synthetic inhibitors and substrates. Protein Sci. 1992, 8, 1007–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, W.S. Molecular biology of plasminogen activators: What are the clinical implications of drug design. Am. J. Cardiol. 1996, 78, 2–7. [Google Scholar]
- Noble, S.; McTavish, D. Reteplase: A review of its pharmacological properties and clinical efficacy in the management of acute myocardial infarction. Drugs 1996, 52, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, P.; Gao, J.; Liao, J.; Sun, S.; Shen, Z.; Qin, S. Recombinant expression of rt-PA gene (encoding Reteplase) in gametophytes of the seaweed Laminaria japonica (Laminariales, Phaeophyta). Sci. China Ser. C Life Sci. 2008, 51, 1116–1120. [Google Scholar] [CrossRef]
- Harris, T.J.; Patel, T.; Marston, F.A.; Little, S.; Emtage, J.S.; Opdenakker, G.; Volckaert, G.; Rombauts, W.; Billiau, A.; De Somer, P. Cloning of cDNA coding for human tissue-type plasminogen activator and its expression in Escherichia coli. Mol. Biol. Med. 1986, 3, 279–292. [Google Scholar]
- Liao, J.; Zhang, J.; Shen, Z. Cloning and expression of tissue-type plasminogen activator mutant Reteplase (r-PA) in E. coli (in Chinese with English abstract). Pharma Biotechnol. 2002, 9, 95–98. [Google Scholar]
- Liu, Y.; Chen, Z.; Lu, H.; Liu, C.; Jin, M.; Guo, Z.; Zhang, W. Optimization of culture medium and photosynthetic characteristics of Platymonas subcordiformis. Chin. J. Process Eng. 2007, 7, 1197–1201. [Google Scholar]
- Xie, J.; Zhang, Y.; Li, Y.; Wang, Y. Mixotrophic cultivation of Platymonas subcordiformis. J. Appl. Phycol. 2001, 13, 343–347. [Google Scholar] [CrossRef]
- Han, P.; Lu, Q.; Fan, L.; Zhou, W. A review on the use of microalgae for sustainable aquaculture. Appl. Sci. 2019, 9, 2377. [Google Scholar] [CrossRef] [Green Version]
- Shi, Q.; Chen, C.; Zhang, W.; Wu, P.; Sun, M.; Wu, H.; Wu, H.; Fu, P.; Fan, J. Transgenic eukaryotic microalgae as green factories: Providing new ideas for the production of biologically active substances. J. Appl. Phycol. 2021, 33, 705–728. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar]




| Samples | Biomass (Fresh Weight) | Soluble Protein | rt-PA | Concentration (μg·mg−1 Soluble Proteins) |
|---|---|---|---|---|
| A1 | 1.93 g | 23.2 mg | 44.2 μg | 1.91 |
| A2 | 2.20 g | 24.8 mg | 20.4 μg | 0.82 |
| B3 | 2.34 g | 23.6 mg | 17.85 μg | 0.76 |
| R1B2 | 1.65 g | 23.6 mg | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.; Zheng, C.; Wang, J.; Jiang, P. Recombinant Expression of Thrombolytic Agent Reteplase in Marine Microalga Tetraselmis subcordiformis (Chlorodendrales, Chlorophyta). Mar. Drugs 2021, 19, 315. https://0-doi-org.brum.beds.ac.uk/10.3390/md19060315
Wu C, Zheng C, Wang J, Jiang P. Recombinant Expression of Thrombolytic Agent Reteplase in Marine Microalga Tetraselmis subcordiformis (Chlorodendrales, Chlorophyta). Marine Drugs. 2021; 19(6):315. https://0-doi-org.brum.beds.ac.uk/10.3390/md19060315
Chicago/Turabian StyleWu, Chunhui, Caiyun Zheng, Jinxia Wang, and Peng Jiang. 2021. "Recombinant Expression of Thrombolytic Agent Reteplase in Marine Microalga Tetraselmis subcordiformis (Chlorodendrales, Chlorophyta)" Marine Drugs 19, no. 6: 315. https://0-doi-org.brum.beds.ac.uk/10.3390/md19060315