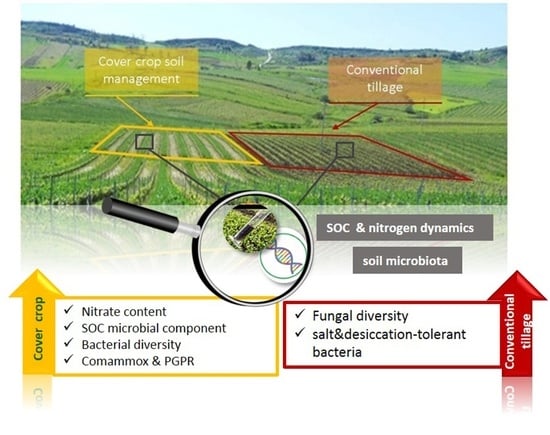
Cover Crop Impact on Soil Organic Carbon, Nitrogen Dynamics and Microbial Diversity in a Mediterranean Semiarid Vineyard
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Soil Analyses
2.3. Incubation Experiment
2.4. DNA Extraction
2.5. Denaturing Gradient Gel Electrophoresis Analysis
2.6. PCR-Amplification of Bacterial and Fungal Ribosomal Intergenic Spacer
2.7. Automated Ribosomal Intergenic Spacer Analysis (ARISA)
2.8. Next Generation Sequencing of Ribosomal DNA and Bioinformatic Analysis
2.9. Statistical Analysis
3. Results
3.1. Soil Organic Carbon and Nitrate Content
3.2. Microbial Biomass and Soil Respiration
3.3. Soil Microbial Diversity
3.4. Diversity and Phylogenetic Composition of CC and CT Soil Bacterial Communities
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pe’er, G.; Bonn, A.; Bruelheide, H.; Dieker, P.; Eisenhauer, N.; Feindt, P.H.; Hagedorn, G.; Hansjürgens, B.; Herzon, I.; Lomba, Â.; et al. Action needed for the EU Common Agricultural Policy to address sustainability challenges. People Nat. 2019. [Google Scholar] [CrossRef]
- Garcia, L.; Celette, F.; Gary, C.; Ripoche, A.; Valdés-Gómez, H.; Metay, A. Management of service crops for the provision of ecosystem services in vineyards: A review. Agric. Ecosyst. Environ. 2018, 251, 158–170. [Google Scholar] [CrossRef] [Green Version]
- Novara, A.; Cerdà, A.; Gristina, L. Sustainable vineyard floor management: An equilibrium between water consumption and soil conservation. Curr. Opin. Environ. Sci. Health 2018, 5, 33–37. [Google Scholar] [CrossRef]
- López-Vicente, M.; García-Ruiz, R.; Guzmán, G.; Vicente-Vicente, J.L.; Van Wesemael, B.; Gómez, J.A. Temporal stability and patterns of runoff and runon with different cover crops in an olive orchard (SW Andalusia, Spain). Catena 2016, 147, 125–137. [Google Scholar] [CrossRef] [Green Version]
- Baiamonte, G.; Minacapilli, M.; Novara, A.; Gristina, L. Time scale effects and interactions of rainfall erosivity and cover management factors on vineyard soil loss erosion in the semi-arid area of southern Sicily. Water 2019, 11, 978. [Google Scholar] [CrossRef] [Green Version]
- García-Díaz, A.; Bienes, R.; Sastre, B.; Novara, A.; Gristina, L.; Cerdà, A. Nitrogen losses in vineyards under different types of soil groundcover. A field runoff simulator approach in central Spain Agriculture. Ecosyst. Environ. 2017, 236, 256–267. [Google Scholar] [CrossRef]
- Peregrina, F.; Pérez-Álvarez, E.P.; Colina, M.; García-Escudero, E. Cover crops and tillage influence soil organic matter and nitrogen availability in a semi-arid vineyard. Arch. Agron. Soil Sci. 2012, 58, 95–102. [Google Scholar] [CrossRef]
- García-González, I.; Hontoria, C.; Gabriel, J.L.; Alonso-Ayuso, M.; Quemada, M. Cover crops to mitigate soil degradation and enhance soil functionality in irrigated land. Geoderma 2018, 322, 81–88. [Google Scholar] [CrossRef]
- Ingels, C.A.; Scow, K.M.; Whisson, D.A.; Drenovsky, R.E. Effects of cover crops on grapevines, yield, juice composition, soil microbial ecology, and gopher activity. Am. J. Enol. Vitic. 2005, 56, 19–29. [Google Scholar]
- Muscas, E.; Cocco, A.; Mercenaro, L.; Cabras, M.; Lentini, A.; Porqueddu, C.; Nieddu, G. Effects of vineyard floor cover crops on grapevine vigor, yield, and fruit quality, and the development of the vine mealybug under a Mediterranean climate Agriculture. Ecosyst. Environ. 2017, 237, 203–212. [Google Scholar] [CrossRef]
- Capó-Bauçà, S.; Marqués, A.; Llopis-Vidal, N.; Bota, J.; Baraza, E. Long-term establishment of natural green cover provides agroecosystem services by improving soil quality in a Mediterranean vineyard. Ecol. Eng. 2019, 127, 285–291. [Google Scholar] [CrossRef]
- Sanz-Cobena, A.; García-Marco, S.; Quemada, M.; Gabriel, J.L.; Almendros, P.; Vallejo, A. Do cover crops enhance N2O, CO2 or CH4 emissions from soil in Mediterranean arable systems? Sci. Total Environ. 2014, 466, 164–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, M.E.; Benitez, E.; Garcia, P.A.; Robles, A.B. Cover crops under different managements vs. frequent tillage in almond orchards in semiarid conditions: Effects on soil quality. Appl. Soil Ecol. 2010, 44, 6–14. [Google Scholar] [CrossRef]
- Ruiz-Colmenero, M.; Bienes, R.; Eldridge, D.J.; Marques, M.J. Vegetation cover reduces erosion and enhances soil organic carbon in a vineyard in the central Spain. Catena 2013, 104, 153–160. [Google Scholar] [CrossRef]
- Morlat, R.; Jacquet, A. Grapevine root system and soil characteristics in a vineyard maintained long-term with or without interrow sward. Am. J. Enol. Vitic. 2003, 54, 1–7. [Google Scholar]
- Celette, F.; Findeling, A.; Gary, C. Competition for nitrogen in an unfertilized intercropping system: The case of an association of grapevine and grass cover in a Mediterranean climate. Eur. J. Agron. 2009, 30, 41–51. [Google Scholar] [CrossRef]
- Laudicina, V.A.; Barbera, V.; Gristina, L.; Badalucco, L. Management practices to preserve soil organic matter in semiarid Mediterranean environments. In Soil Organic Matter: Ecology, Environmental Impact and Management; Björklund, P.A., Mello, F.V., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2012; pp. 39–61. [Google Scholar]
- Unkovich, M.J.; Pate, J. An appraisal of recent field measurements of symbiotic N2 fixation by annual legumes. Field Crops Res. 2000, 65, 211–228. [Google Scholar] [CrossRef]
- Romdhane, S.; Spor, A.; Busset, H.; Falchetto, L.; Martin, J.; Bizouard, F.; Bru, D.; Breuil, M.C.; Philippot, L.; Cordeau, S. Cover Crop Management Practices Rather than Composition of Cover Crop Mixtures Affect Bacterial Communities in No-Till Agroecosystems. Front. Microbiol. 2019, 10, 1618. [Google Scholar] [CrossRef]
- Lopes, C.M.; Monteiro, A.; Machado, J.P.; Fernandes, N.; Araújo, A. Cover cropping in a slopping non-irrigated vineyard: II—Effects on vegetative growth, yield, berry and wine quality of ‘Cabernet Sauvignon’ grapevines. Ciência Téc. Vitiv 2008, 23, 37–43. [Google Scholar]
- Delpuech, X.; Metay, A. Adapting cover crop soil coverage to soil depth to limit competition for water in a Mediterranean vineyard. Eur. J. Agron. 2018, 97, 60–69. [Google Scholar] [CrossRef]
- Novara, A.; Gristina, L.; Saladino, S.S.; Santoro, A.; Cerdà, A. Soil erosion assessment on tillage and alternative soil managements in a Sicilian vineyard. Soil Tillage Res. 2011, 117, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Finney, D.M.; Buyer, J.S.; Kaye, J.P. Living cover crops have immediate impacts on soil microbial community structure and function. J. Soil Water Conserv. 2017, 724, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Strickland, M.S.; Thomason, W.E.; Avera, B.; Franklin, J.; Minick, K.; Yamada, S.; Badgley, B.D. Short-Term Effects of Cover Crops on Soil Microbial Characteristics and Biogeochemical Processes across Actively Managed Farms. Agrosyst. Geosci. Environ. 2019, 2. [Google Scholar] [CrossRef] [Green Version]
- WRB 2006. IUSS Working Group World Reference Base for Soil Resources 2006; World Soil Resources Reports No. 103; FAO: Rome, Italy, 2006; ISBN 92-5-105511-4. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties, 2nd ed.; Page, A.L., Ed.; Agronomy Series No. 9, ASA; SSSA: Madison, WI, USA, 1996. [Google Scholar]
- Catania, V.; Santisi, S.; Signa, G.; Vizzini, S.; Mazzola, A.; Cappello, S.; Yakimov, M.M.; Quatrini, P. Intrinsic bioremediation potential of a chronically polluted marine coastal area. Mar. Pollut. Bull. 2015, 99, 138–149. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.; Uittierlinden, A.G. Profiling of Complex Microbial Populations by Denaturing Gradient Gel Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes Coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Catania, V.; Sarà, G.; Settanni, L.; Quatrini, P. Bacterial communities in sediment of a Mediterranean marine protected area. Can. J. Microbiol. 2016, 63, 303–311. [Google Scholar] [CrossRef]
- Catania, V.; Cappello, S.; Di Giorgi, V.; Santisi, S.; Di Maria, R.; Mazzola, A. Microbial communities of polluted sub-surface marine sediments. Mar. Pollut. Bull. 2018, 131, 396–406. [Google Scholar] [CrossRef]
- Djahnit, N.; Chernai, S.; Catania, V.; Hamdi, B.; China, B.; Cappello, S.; Quatrini, P. Isolation, characterization and determination of biotechnological potential of oil-degrading bacteria from Algerian centre coast. J. Appl. Microbiol. 2019, 126, 780–795. [Google Scholar] [CrossRef]
- Cardinale, M.; Brusetti, L.; Quatrini, P.; Borin, S.; Puglia, A.M.; Rizzi, A.; Zanardini, E.; Sorlini, C.; Corselli, C.; Daffonchio, D. Comparison of Different Primer Sets for Use in Automated Ribosomal Intergenic Spacer Analysis of Complex Bacterial Communities. Appl. Environ. Microbiol. 2004, 70, 6147–6156. [Google Scholar] [CrossRef] [Green Version]
- Ranjard, L.; Poly, F.; Lata, J.C.; Mougel, C.; Thioulouse, J.; Nazaret, S. Characterization of Bacterial and Fungal Soil Communities by Automated Ribosomal Intergenic Spacer Analysis Fingerprints: Biological and Methodological Variability. Appl. Environ. Microbiol. 2001, 67, 4479–4487. [Google Scholar] [CrossRef] [Green Version]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 182, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Slabbert, E.; Kongor, R.Y.; Esler, K.J.; Jacobs, K. Microbial diversity and community structure in Fynbos soil. Mol. Ecol. 2010, 19, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 98, e105592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 13 November 2019).
- Bushnell, B. BBMap: A Fast. Accurate. Splice-aware Aligner No. LBNL-7065E; Lawrence Berkeley National Lab. (LBNL): Berkeley, CA, USA, 2014.
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 2514, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information NCBI Resource Coordinators. Nucleic Acids Res. 2016, 44, D7. [Google Scholar] [CrossRef] [Green Version]
- Mcmurdie, P.J.; Holmes, S.; Watson, M. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 84, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SPSS Inc. PASW Statistics for Windows; Version 18.0.; SPSS Inc.: Chicago, IL, USA, 2009. [Google Scholar]
- Marstorp, H.; Guan, X.; Gong, P. Relationship between dsDNA, chloroform labile C and ergosterol in soils of different organic matter contents and pH. Soil Biol. Biochem. 2000, 326, 879–882. [Google Scholar] [CrossRef]
- Anderson, T.H. Microbial Eco-Physiological Indicators to Asses Soil Quality. Agric. Ecosyst. Environ. 2003, 98, 285–293. [Google Scholar] [CrossRef]
- Vukicevich, E.; Lowery, T.; Bowen, P.; Úrbez-Torres, J.R.; Hart, M. Cover crops to increase soil microbial diversity and mitigate decline in perennial agriculture. A review. Agron. Sustain. Dev. 2016, 363, 48. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef] [Green Version]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, B.; Winsley, T.; Ji, M.; Neilan, B. Insights into the distribution and abundance of the ubiquitous candidatus Saccharibacteria phylum following tag pyrosequencing. Sci. Rep. 2014, 41, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taubert, M.; Stähly, J.; Kolb, S.; Küsel, K. Divergent microbial communities in groundwater and overlying soils exhibit functional redundancy for plant-polysaccharide degradation. PLoS ONE 2019, 143, e0212937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Liu, S. Insight into the overconsumption of ammonium by anammox consortia under anaerobic conditions. J. Appl. Microbial. 2014, 1176, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Mohammadipanah, F.; Wink, J. Actinobacteria from Arid and Desert Habitats: Diversity and Biological Activity. Front. Microbiol. 2016, 6, 1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, A.J.; Bowyer, J.; Holley, M.P.; O’Donoghue, M.; Montgomery, M.; Gillings, M.R. Diverse.; yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 2000, 332, 111–120. [Google Scholar] [CrossRef]
- Pascual, J.; Wüst, P.K.; Geppert, A.; Foesel, B.U.; Huber, K.J.; Overmann, J. Novel isolates double the number of chemotrophic species and allow the first description of higher taxa in Acidobacteria subdivision 4. Syst. Appl. Microbiol. 2015, 388, 534–544. [Google Scholar] [CrossRef] [PubMed]
- De Jesus Sousa, J.A.; Olivares, F.L. Plant growth promotion by streptomycetes: Ecophysiology, mechanisms and applications. Chem. Biol. Technol. Agric. 2016, 31, 24. [Google Scholar] [CrossRef] [Green Version]
- García-Díaz, A.; Allas, R.B.; Gristina, L.; Cerdà, A.; Pereira, P.; Novara, A. Carbon input threshold for soil carbon budget optimization in eroding vineyards. Geoderma 2016, 271, 144–149. [Google Scholar] [CrossRef] [Green Version]
- Novara, A.; Minacapilli, M.; Santoro, A.; Rodrigo-Comino, J.; Carrubba, A.; Sarno, M.; Venezia, G.; Gristina, L. Real cover crops contribution to soil organic carbon sequestration in sloping vineyard. Sci. Total Environ. 2019, 652, 300–306. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, A.L.; Sheaffer, C.C.; Wyse, D.L.; Staley, C.; Gould, T.J.; Sadowsky, M.J. Associations between soil bacterial community structure and nutrient cycling functions in long-term organic farm soils following cover crop and organic fertilizer amendment. Sci. Total. Environ. 2016, 566, 949–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrera, L.M.; Buyer, J.S.; Vinyard, B.; Abdul-Baki, A.A.; Sikora, L.J.; Teasdale, J.R. Effects of cover crops. compost. and manure amendments on soil microbial community structure in tomato production systems. Appl. Soil Ecol. 2007, 37, 247–255. [Google Scholar] [CrossRef]
- Buyer, J.S.; Teasdale, J.R.; Roberts, D.P.; Zasada, I.A.; Maul, J.E. Factors affecting soil microbial community structure in tomato cropping systems. Soil Biol. Biochem. 2010, 42, 831–841. [Google Scholar] [CrossRef]
- Kallenbach, C.; Frey, S.; Grandy, A. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 2016, 7, 13630. [Google Scholar] [CrossRef]
- Schmidt, R.; Gravuer, K.; Bossange, A.V.; Mitchell, J.; Scow, K. Long-term use of cover crops and no-till shift soil microbial community life strategies in agricultural soil. PLoS ONE 2018. [Google Scholar] [CrossRef]
- Bonanomi, G.; De Filippis, F.; Cesarano, G.; La Storia, A.; Ercolini, D.; Scala, F. Organic farming induces changes in soil microbiota that affect agro-ecosystem functions. Soil Biol. Biochem. 2016, 103, 327–336. [Google Scholar] [CrossRef]
- Beeckman, F.; Motte, H.; Beeckman, T. Nitrification in agricultural soils: Impact, actors and mitigation. Curr. Opin. Biotechnol. 2018, 50, 166–173. [Google Scholar] [CrossRef]
- IPCC. Working Group I Contribution to the IPCC Fifth Assessment Report: Climate Change 2013: The Physical Science Basis; Summary for Policymakers; IPCC: Geneva, Switzerland, 2013. [Google Scholar]
- Zhou, Y.; Zhu, H.; Fu, S.; Yao, Q. Variation in soil microbial community structure associated with different legume species is greater than that associated with different grass species. Front. Microbial. 2017, 31, 1007. [Google Scholar] [CrossRef]
- Orellana, L.H.; Chee-Sanford, J.C.; Sanford, R.A.; Löffler, F.E.; Konstantinidis, K.T. Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl. Environ. Microbiol. 2018, 84, e01646-17. [Google Scholar] [CrossRef] [Green Version]
- Subhash, Y.; Tushar, L.; Sasikala, C.; Ramana, C.V. Erythrobacter odishensis sp. nov. and Pontibacter odishensis sp. nov. isolated from dry soil of a solar saltern. Int. J. Syst. Evol. Microbiol. 2013, 6312, 4524–4532. [Google Scholar] [CrossRef]
- De Luca, G.; Barakat, M.; Ortet, P.; Fochesato, S.; Jourlin-Castelli, C.; Ansaldi, M.; Bastien, O. The cyst-dividing bacterium Ramlibacter tataouinensis TTB310 genome reveals a well-stocked toolbox for adaptation to a desert environment. PLoS ONE 2011, 69, e23784. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Błaszczyk, M. Indicators of arable soils fatigue–Bacterial families and genera: A metagenomic approach. Ecol. Indic. 2018, 93, 490–500. [Google Scholar] [CrossRef]
- Shao, M.F.; Zhang, T.; Fang, H.H.P. Sulfur-driven autotrophic denitrification: Diversity; biochemistry; and engineering applications. Appl. Microbiol. Biotechnol. 2010, 885, 1027–1042. [Google Scholar] [CrossRef]
- ECA. European Court of Auditors Special Report n°33/2018: Combating Desertification in the EU: A Growing Threat in Need of More Action; ECA: Luxembourg, 2018. [Google Scholar]







| CT | CC16 | CC17 | |
|---|---|---|---|
| Total microbial biomass (µg DNAg−1 soil DW) | 6.59b | 12.72a | 11.87a |
| Extractable organic C (mg kg−1) | 21.22a | 17.99b | 20.32a |
| Biological available C (mg CO2-C kg−1) | # | 41.76a | 43.23b |
| k | # | 0.001b | 0.003a |
| MRT (1/k) | # | 1000a | 333b |
| Total mineralized C (mg CO2-C kg−1) | 1028b | 1025b | 1034a |
| Index | Bacteria (B-ARISA) | Fungi (F-ARISA) | ||||
|---|---|---|---|---|---|---|
| CC16 | CC17 | CT | CC16 | CC17 | CT | |
| Taxa_S | 17 | 14 | 11 | 18 | 22 | 29 |
| Individuals | 4458 | 4069 | 3798 | 4597 | 4287 | 4751 |
| Dominance_D | 0.31 | 0.40 | 0.38 | 0.26 | 0.20 | 0.14 |
| Simpson_1-D | 0.70 | 0.60 | 0.62 | 0.74 | 0.80 | 0.86 |
| Shannon_H | 1.86 | 1.55 | 1.55 | 2.05 | 2.33 | 2.71 |
| Evenness_e^H/S | 0.38 | 0.34 | 0.43 | 0.43 | 0.47 | 0.52 |
| Brillouin | 1.84 | 1.54 | 1.54 | 2.04 | 2.32 | 2.69 |
| Menhinick | 0.25 | 0.22 | 0.18 | 0.27 | 0.34 | 0.42 |
| Margalef | 1.90 | 1.56 | 1.21 | 2.02 | 2.51 | 3.31 |
| Equitability_J | 0.66 | 0.59 | 0.64 | 0.71 | 0.75 | 0.80 |
| Fisher_alpha | 2.24 | 1.81 | 1.39 | 2.38 | 3.03 | 4.11 |
| Berger-Parker | 0.53 | 0.62 | 0.60 | 0.48 | 0.42 | 0.35 |
| Alpha Diversity | Phylum | Class | Order | Family | Genus | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC17 | CT | CC17 | CT | CC17 | CT | CC17 | CT | CC17 | CT | |
| Observed | 25 | 25 | 53 | 53 | 91 | 92 | 138 | 140 | 263 | 263 |
| Chao1 | 25 | 25 | 53 | 53 | 91 | 92 | 138 | 140 | 263 | 263 |
| Shannon | 2.19 | 2.16 | 2.93 | 2.89 | 3.02 | 3.01 | 2.90 | 2.91 | 2.73 | 2.69 |
| Simpson | 0.85 | 0.84 | 0.92 | 0.91 | 0.88 | 0.88 | 0.80 | 0.81 | 0.70 | 0.71 |
| Fisher | 2.56 | 2.40 | 5.95 | 5.53 | 10.97 | 10.25 | 17.64 | 16.47 | 37.15 | 33.80 |
| Bacterial Genera | Abundance (Reads %) | Abundance Ratio | ||
|---|---|---|---|---|
| CT | CC17 | CC17/CT | CT17/CC | |
| Stenotrophobacter | 1.16 | 1.76 | 1.52 | |
| Nitrospira | 0.57 | 0.91 | 1.61 | |
| Streptomyces | 0.39 | 0.66 | 1.68 | |
| Microvirga | 1.71 | 0.99 | 1.72 | |
| Massilia | 1.07 | 0.40 | 2.69 | |
| Ramlibacter | 0.80 | 0.50 | 1.58 | |
| Pseudarthrobacter | 0.78 | 0.34 | 2.33 | |
| Pontibacter | 0.59 | 0.29 | 2.02 | |
| Thiobacillus | 0.54 | 0.02 | 23.77 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novara, A.; Catania, V.; Tolone, M.; Gristina, L.; Laudicina, V.A.; Quatrini, P. Cover Crop Impact on Soil Organic Carbon, Nitrogen Dynamics and Microbial Diversity in a Mediterranean Semiarid Vineyard. Sustainability 2020, 12, 3256. https://0-doi-org.brum.beds.ac.uk/10.3390/su12083256
Novara A, Catania V, Tolone M, Gristina L, Laudicina VA, Quatrini P. Cover Crop Impact on Soil Organic Carbon, Nitrogen Dynamics and Microbial Diversity in a Mediterranean Semiarid Vineyard. Sustainability. 2020; 12(8):3256. https://0-doi-org.brum.beds.ac.uk/10.3390/su12083256
Chicago/Turabian StyleNovara, Agata, Valentina Catania, Marco Tolone, Luciano Gristina, Vito Armando Laudicina, and Paola Quatrini. 2020. "Cover Crop Impact on Soil Organic Carbon, Nitrogen Dynamics and Microbial Diversity in a Mediterranean Semiarid Vineyard" Sustainability 12, no. 8: 3256. https://0-doi-org.brum.beds.ac.uk/10.3390/su12083256