Growth and Toxin Production of Gambierdiscus spp. Can Be Regulated by Quorum-Sensing Bacteria
Abstract
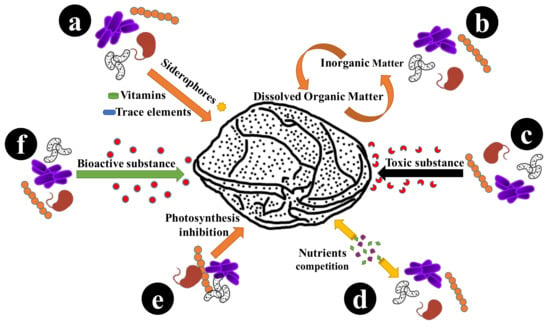
:1. Introduction
2. Results
2.1. QS Bacteria Screening
2.2. Effects of QS Bacteria on Algal Growth
2.3. The Growth Promotion Principle of QS Bacteria
2.4. Influence of QS Bacteria on Algal Toxicity
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection and Algal Culture
5.2. Bacterial Isolation and AHL-Producer Identification
5.3. Co-Culture Experiment at Early Log Phase
5.4. Co-Culture Experiment at Stationary Phase and Decline Phase
5.5. Bioactive Substance Investigation
5.6. Species-Specificity Survey
5.7. Algal Toxicity Study
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grattan, L.M.; Holobaugh, S.; Morris, J.G. Harmful algal blooms and public health. Harmful Algae 2016, 70, 859–875. [Google Scholar] [CrossRef] [PubMed]
- Chinain, M.; Darius, H.T.; Ung, A.; Fouc, M.T.; Revel, T.; Cruchet, P.; Pauillac, S.; Laurent, D. Ciguatera risk management in French Polynesia: The case study of Raivavae Island (Australes Archipelago). Toxicon 2010, 56, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Chinain, M.; Darius, H.T.; Ung, A.; Cruchet, P.; Wang, Z.; Ponton, D.; Laurent, D.; Pauillac, S. Growth and toxin production in the ciguatera-causing dinoflagellate Gambierdiscus polynesiensis (Dinophyceae) in culture. Toxicon 2010, 56, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J. The changing face of ciguatera. Toxicon 2001, 39, 97–106. [Google Scholar] [CrossRef]
- Hamilton, B.; Whittle, N.; Shaw, G.; Eaglesham, G.; Moore, M.R.; Lewis, R.J. Human fatality associated with Pacific ciguatoxin contaminated fish. Toxicon 2010, 56, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Hung, P.; Lo, J.Y. Ciguatera fish poisoning in Hong Kong–a 10-year perspective on the class of ciguatoxins. Toxicon 2014, 86, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.J. Gambierdiscus yasumotoi sp. nov. (Dinophyceae), a toxic benthic dinoflagellate from southeastern Asia. J. Phycol. 1998, 34, 661–668. [Google Scholar] [CrossRef]
- Lewis, R.J. Ciguatera: Australian perspectives on a global problem. Toxicon 2006, 48, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Barton, E.D.; Tanner, P.; Turchen, S.G.; Tunget, C.L.; Manoguerra, A.; Clark, R.F. Ciguatera fish poisoning. A southern California epidemic. West. J. Med. 1995, 163, 31. [Google Scholar] [PubMed]
- Van Dolah, F.M. Marine algal toxins: Origins, health effects, and their increased occurrence. Environ. Health Perspect. 2000, 108 (Suppl. 1), 133. [Google Scholar] [CrossRef]
- Caillaud, A.; De la Iglesia, P.; Darius, H.T.; Pauillac, S.; Aligizaki, K.; Fraga, S.; Chinain, M.; Diogène, J. Update on methodologies available for ciguatoxin determination: Perspectives to confront the onset of ciguatera fish poisoning in Europe. Mar. Drugs 2010, 8, 1838–1907. [Google Scholar] [CrossRef] [PubMed]
- Rongo, T.; van Woesik, R. Socioeconomic consequences of ciguatera poisoning in Rarotonga, southern Cook Islands. Harmful Algae 2012, 20, 92–100. [Google Scholar] [CrossRef]
- Yogi, K.; Oshiro, N.; Inafuku, Y.; Hirama, M.; Yasumoto, T. Detailed LC-MS/MS analysis of ciguatoxins revealing distinct regional and species characteristics in fish and causative alga from the Pacific. Anal. Chem. 2011, 83, 8886–8891. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Luo, Q.; Liang, Y.; Mazumder, A. Processes and pathways of ciguatoxin in aquatic food webs and fish poisoning of seafood consumers. Environ. Rev. 2016, 24, 144–150. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Rivera, E.; Villareal, T.A. Macroalgal palatability and the flux of ciguatera toxins through marine food webs. Harmful Algae 2006, 5, 497–525. [Google Scholar] [CrossRef]
- Yasumoto, T. The chemistry and biological function of natural marine toxins. Chem. Rec. 2001, 1, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.H.; Mak, Y.L.; Wu, J.J.; Jin, L.; Sit, W.H.; Lam, J.C.W.; Mitcheson, Y.S.; Chan, L.L.; Lam, P.K.S.; Murphy, M.B. Spatial distribution of ciguateric fish in the Republic of Kiribati. Chemosphere 2011, 84, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Mak, Y.L.; Wai, T.C.; Murphy, M.B.; Chan, W.H.; Wu, J.J.; Lam, J.C.W.; Chan, L.L.; Lam, P.K.S. Pacific ciguatoxins in food web components of coral reef systems in the Republic of Kiribati. Environ. Sci. Technol. 2013, 47, 14070–14079. [Google Scholar] [CrossRef] [PubMed]
- Mak, Y.L. Pacific-Ciguatoxins (P-CTXs) in Coral Reef Fishes: Toxin Purification, Analytical Method Validation and Trophodynamics in Marine Food Web. Ph.D. Thesis, City University of Hong Kong, Hong Kong, China, 2012. [Google Scholar]
- Ashton, M.; Rosado, W.; Govind, N.S.; Tosteson, T.R. Culturable and nonculturable bacterial symbionts in the toxic benthic dinoflagellate Ostreopsis lenticularis. Toxicon 2003, 42, 419–424. [Google Scholar] [CrossRef]
- Lartigue, J.; Jester, E.L.; Dickey, R.W.; Villareal, T.A. Nitrogen source effects on the growth and toxicity of two strains of the ciguatera-causing dinoflagellate Gambierdiscus toxicus. Harmful Algae 2009, 8, 781–791. [Google Scholar] [CrossRef]
- Parsons, M.L.; Settlemier, C.J.; Bienfang, P.K. A simple model capable of simulating the population dynamics of Gambierdiscus, the benthic dinoflagellate responsible for ciguatera fish poisoning. Harmful Algae 2010, 10, 71–80. [Google Scholar] [CrossRef]
- Kibler, S.R.; Litaker, R.W.; Holland, W.C.; Vandersea, M.W.; Tester, P.A. Growth of eight Gambierdiscus (Dinophyceae) species: Effects of temperature, salinity and irradiance. Harmful Algae 2012, 19, 1–14. [Google Scholar] [CrossRef]
- Sakami, T.; Nakahara, H.; Chinain, M.; Ishida, Y. Effects of epiphytic bacteria on the growth of the toxic dinoflagellate Gambierdiscus toxicus (Dinophyceae). J. Exper. Mar. Biol. Ecol. 1999, 233, 231–246. [Google Scholar] [CrossRef]
- Burke, C.; Thomas, T.; Lewis, M.; Steinberg, P.; Kjelleberg, S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011, 5, 590. [Google Scholar] [CrossRef] [PubMed]
- Rains, L.K.; Parsons, M.L. Gambierdiscus species exhibit different epiphytic behaviors toward a variety of macroalgal hosts. Harmful Algae 2015, 49, 29–39. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, G.; Zhu, X.; Chen, L.; Cai, Z. A review of the relationship between algae and bacteria in harmful algal blooms. Acta Ecol. Sin. 2014, 34, 269–281. [Google Scholar] [CrossRef]
- Zhou, J.; Lyu, Y.; Richlen, M.L.; Anderson, D.M.; Cai, Z. Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. Crit. Rev. Plant Sci. 2016, 35, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Ashen, J.B.; Goff, L.J. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl. Environ. Microbiol. 2000, 66, 3024–3030. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- Luc Rolland, J.; Stien, D.; Sanchez-Ferandin, S.; Lami, R. Quorum sensing and quorum quenching in the phycosphere of phytoplankton: A case of chemical interactions in ecology. J. Chem. Ecol. 2016, 42, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Parsek, M.R.; Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001, 35, 439–468. [Google Scholar] [CrossRef] [PubMed]
- Joint, I.; Tait, K.; Callow, M.E.; Callow, J.A.; Milton, D.; Williams, P.; Cámara, M. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 2002, 298, 1207–1207. [Google Scholar] [CrossRef] [PubMed]
- Tait, K.; Joint, I.; Daykin, M.; Milton, D.L.; Williams, P.; Camara, M. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 2005, 7, 229–240. [Google Scholar] [CrossRef] [PubMed]
- González, J.E.; Keshavan, N.D. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 2006, 70, 859–875. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Reddy, C.R.K. Seaweed–microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 2014, 88, 213–230. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Baghel, R.S.; Reddy, C.R.K.; Jha, B. Effect of quorum sensing signals produced by seaweed-associated bacteria on carpospore liberation from Gracilaria dura. Front. Plant Sci. 2015, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, T.; Yamaguchi, H.; Iwamoto, H.; Nishimura, T.; Adachi, M. Effects of temperature, salinity and their interaction on growth of Japanese Gambierdiscus spp. (Dinophyceae). Harmful Algae 2014, 35, 29–37. [Google Scholar] [CrossRef]
- Lehane, L.; Lewis, R.J. Ciguatera: Recent advances but the risk remains. Int. J. Food Microbiol. 2000, 61, 91–125. [Google Scholar] [CrossRef]
- Chinain, M.; Laurent, N.; Pauillac, S.; Legrand, A.M. Effects of an artificial medium and axenic culturing on toxicity of cultured Gambierdiscus toxicus. In Proceedings of the International Symposium on Ciguatera and Marine Natural Products; Hokama, Y., Scheuer, P.J., Yasumoto, T., Eds.; Asian Pacific Research Foundation: Hawaii, HI, USA, 1994; pp. 91–98. [Google Scholar]
- Soria-Dengg, S.; Reissbrodt, R.A.H.U.; Horstmann, U. Siderophores in marine coastal waters and their relevance for iron uptake by phytoplankton: Experiments with the diatom Phaeodactylum tricornutum. Mar Ecol. Prog. Ser. 2001, 220, 73–82. [Google Scholar] [CrossRef]
- Gutierrez, T.; Biller, D.V.; Shimmield, T.; Green, D.H. Metal binding properties of the EPS produced by Halomonas sp. TG39 and its potential in enhancing trace element bioavailability to eukaryotic phytoplankton. Biometals 2012, 25, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, J.L.; Garbayo, I.; Cuaresma, M.; Montero, Z.; González-del-Valle, M.; Vílchez, C. Impact of microalgae-bacteria interactions on the production of algal biomass and associated compounds. Mar. Drugs 2016, 14, 100. [Google Scholar] [CrossRef] [PubMed]
- Azam, F. Microbial control of oceanic carbon flux: The plot thickens. Science 1998, 280, 694–696. [Google Scholar] [CrossRef]
- Russo, D.A.; Couto, N.; Beckerman, A.P.; Pandhal, J. A Metaproteomic analysis of the response of a freshwater microbial community under nutrient enrichment. Front. Microbiol. 2016, 7, 1172. [Google Scholar] [CrossRef] [PubMed]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Di Padova, F. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarska, I.; Ehrman, J.M.; Bates, S.S.; Green, D.H.; Léger, C.; Harris, J. Diversity and distribution of epibiotic bacteria on Pseudo-nitzschia multiseries (Bacillariophyceae) in culture, and comparison with those on diatoms in native seawater. Harmful Algae 2005, 4, 725–741. [Google Scholar] [CrossRef]
- Xu, Y.; Richlen, M.L.; Morton, S.L.; Mak, Y.L.; Chan, L.L.; Tekiau, A.; Anderson, D.M. Distribution, abundance and diversity of Gambierdiscus spp. from a ciguatera-endemic area in Marakei, Republic of Kiribati. Harmful Algae 2014, 34, 56–68. [Google Scholar] [CrossRef]
- Keller, M.D.; Selvin, R.C.; Claus, W.; Guillard, R.R. Media for the culture of oceanic ultraphytoplankton 1, 2. J. Phycol. 1987, 23, 633–638. [Google Scholar] [CrossRef]
- Radjasa, O.K.; Urakawa, H.; Kita-Tsukamoto, K.; Ohwada, K. Characterization of psychrotrophic bacteria in the surface and deep-sea waters from the northwestern Pacific Ocean based on 16S ribosomal DNA analysis. Mar. Biotechnol. 2001, 3, 454–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Chai, Y.; Zhong, Z.; Li, S.; Winans, S.C. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: Detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 2003, 69, 6949–6953. [Google Scholar] [CrossRef] [PubMed]
- Jakob, T.; Schreiber, U.; Kirchesch, V.; Langner, U.; Wilhelm, C. Estimation of chlorophyll content and daily primary production of the major algal groups by means of multiwavelength-excitation PAM chlorophyll fluorometry: Performance and methodological limits. Photosynth. Res. 2005, 83, 343–361. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Mak, Y.L.; Murphy, M.B.; Lam, J.C.; Chan, W.H.; Wang, M.; Chan, L.L.; Lam, P.K.S. Validation of an accelerated solvent extraction liquid chromatography–tandem mass spectrometry method for Pacific ciguatoxin-1 in fish flesh and comparison with the mouse neuroblastoma assay. Anal. Bioanal. Chem. 2011, 400, 3165–3175. [Google Scholar] [CrossRef] [PubMed]







| No. * | Description | Bit Score | E Value | Ident |
|---|---|---|---|---|
| 1 | Vibrio sp. WC141014 16S ribosomal RNA gene, partial sequence | 1836 | 0 | 99% |
| 2 | Alteromonas macleodii strain KS62 16S ribosomal RNA gene, partial sequence | 2567 | 0 | 99% |
| 3 | Marinobacter hydrocarbonoclasticus strain NIOSSD020#224 16S ribosomal RNA gene, partial sequence | 2097 | 0 | 97% |
| 4 | Thalassospira sp. KU27D2 gene for 16S rRNA, partial sequence | 1611 | 0 | 99% |
| 5 | Pseudomonas aeruginosa strain CFV 16S ribosomal RNA gene, partial sequence | 1094 | 0 | 100% |
| 6 | Vibrio maritimus strain CR-IV-34 16S ribosomal RNA gene, partial sequence | 2084 | 0 | 97% |
| 7 | Providencia vermicola strain NBA-2365 16S ribosomal RNA gene, partial sequence | 1131 | 0 | 99% |
| 8 | Marinobacterium stanieri S30 contig00002, whole genome shotgun sequence | 2615 | 0 | 99% |
| 9 | Bacillus anthracis strain C1E4 16S ribosomal RNA gene, partial sequence | 1962 | 0 | 99% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Yao, M.; Zhou, J.; Tan, S.; Jin, H.; Zhang, F.; Mak, Y.L.; Wu, J.; Lai Chan, L.; Cai, Z. Growth and Toxin Production of Gambierdiscus spp. Can Be Regulated by Quorum-Sensing Bacteria. Toxins 2018, 10, 257. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10070257
Wang B, Yao M, Zhou J, Tan S, Jin H, Zhang F, Mak YL, Wu J, Lai Chan L, Cai Z. Growth and Toxin Production of Gambierdiscus spp. Can Be Regulated by Quorum-Sensing Bacteria. Toxins. 2018; 10(7):257. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10070257
Chicago/Turabian StyleWang, Bo, Mimi Yao, Jin Zhou, Shangjin Tan, Hui Jin, Feng Zhang, Yim Ling Mak, Jiajun Wu, Leo Lai Chan, and Zhonghua Cai. 2018. "Growth and Toxin Production of Gambierdiscus spp. Can Be Regulated by Quorum-Sensing Bacteria" Toxins 10, no. 7: 257. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins10070257