Protein-Lipid Interaction of Cytolytic Toxin Cyt2Aa2 on Model Lipid Bilayers of Erythrocyte Cell Membrane
Abstract
:1. Introduction
2. Results
2.1. Determination of the Cyt2Aa2-Lipid Interaction with Different Model Lipid Bilayers that Mimic the Erythrocyte Membrane by QCM-D
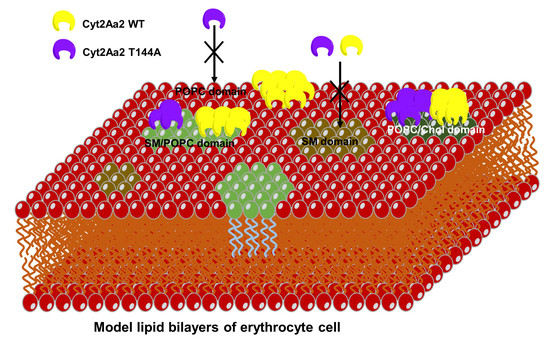
2.2. AFM Imaging of the Cyt2Aa2 (WT and Mutant) Interaction with Different Model Lipid Bilayers
2.3. Cyt2Aa2 (WT and Mutant) Toxin Interaction with Erythrocyte Cell Membranes
3. Discussion
4. Materials and Methods
4.1. Reagents and Buffer
4.2. Protein Preparation
4.3. Lipid Vesicle Preparation
4.4. Supported Erythrocyte Cell Membrane Preparation
4.5. Quartz Crystal Microbalance with Dissipation (QCM-D) Measurement
4.6. Atomic Force Microscopy (AFM) Imaging
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hofte, H.; Whiteley, H.R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 1989, 53, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Angsuthanasombat, C.; Crickmore, N.; Ellar, D.J. Effects on toxicity of eliminating a cleavage site in a predicted interhelical loop in Bacillus thuringiensis CryIVB delta-endotoxin. FEMS Microbiol. Lett. 1993, 111, 255–261. [Google Scholar] [PubMed] [Green Version]
- Crickmore, N.; Bone, E.J.; Williams, J.A.; Ellar, D.J. Contribution of the individual components of the [delta]-endotoxin crystal to the mosquitocidal activity of Bacillus thuringiensis subsp. israelensis. FEMS Microbiol. Lett. 1995, 131, 249–254. [Google Scholar] [CrossRef]
- Al-yahyaee, S.A.S.; Ellar, D.J. Maximal toxicity of cloned CytA δ-endotoxin from Bacillus thuringiensis subsp. israelensis requires proteolytic processing from both the N- and C-termini. Microbiology 1995, 141, 3141–3148. [Google Scholar] [CrossRef] [Green Version]
- Butko, P. Cytolytic toxin Cyt1A and its mechanism of membrane damage: Data and hypotheses. Appl. Environ. Microbiol. 2003, 69, 2415–2422. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Lopez, L.; Soberon, M.; Bravo, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 2013, 37, 3–22. [Google Scholar] [CrossRef] [Green Version]
- Thomas, W.E.; Ellar, D.J. Bacillus thuringiensis var israelensis crystal delta-endotoxin: Effects on insect and mammalian cells in vitro and in vivo. J. Cell Sci. 1983, 60, 181–197. [Google Scholar]
- Gill, S.S.; Cowles, E.A.; Pietrantonio, P.V. The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 1992, 37, 615–636. [Google Scholar] [CrossRef]
- Pigott, C.R.; Ellar, D.J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 2007, 71, 255–281. [Google Scholar] [CrossRef] [Green Version]
- Thomas, W.E.; Ellar, D.J. Mechanism of action of Bacillus thuringiensis var israelensis insecticidal delta-endotoxin. FEBS Lett. 1983, 154, 362–368. [Google Scholar] [CrossRef] [Green Version]
- Ben-Dov, E. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins (Basel) 2014, 6, 1222–1243. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.L.; Soccol, V.T.; Soccol, C.R. Bacillus thuringiensis: Mechanism of action, resistance, and new applications: A review. Crit. Rev. Biotechnol. 2016, 36, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.; Pardo-Lopez, L.; Munoz-Garay, C.; Fernandez, L.E.; Perez, C.; Sanchez, J.; Soberon, M.; Bravo, A. Role of receptor interaction in the mode of action of insecticidal Cry and Cyt toxins produced by Bacillus thuringiensis. Peptides 2007, 28, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Promdonkoy, B.; Chewawiwat, N.; Tanapongpipat, S.; Luxananil, P.; Panyim, S. Cloning and characterization of a cytolytic and mosquito larvicidal delta-endotoxin from Bacillus thuringiensis subsp. darmstadiensis. Curr. Microbiol. 2003, 46, 94–98. [Google Scholar] [CrossRef]
- Cahan, R.; Friman, H.; Nitzan, Y. Antibacterial activity of Cyt1Aa from Bacillus thuringiensis subsp. israelensis. Microbiology 2008, 154, 3529–3536. [Google Scholar] [CrossRef] [Green Version]
- Knowles, B.H.; Blatt, M.R.; Tester, M.; Horsnell, J.M.; Carroll, J.; Menestrina, G.; Ellar, D.J. A cytolytic delta-endotoxin from Bacillus thuringiensis var. israelensis forms cation-selective channels in planar lipid bilayers. FEBS Lett. 1989, 244, 259–262. [Google Scholar] [CrossRef] [Green Version]
- Promdonkoy, B.; Ellar, D.J. Investigation of the pore-forming mechanism of a cytolytic delta-endotoxin from Bacillus thuringiensis. Biochem. J. 2003, 374, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Butko, P.; Huang, F.; Pusztai-Carey, M.; Surewicz, W.K. Membrane permeabilization induced by cytolytic delta-endotoxin CytA from Bacillus thuringiensis var. israelensis. Biochemistry 1996, 35, 11355–11360. [Google Scholar] [CrossRef]
- Manceva, S.D.; Pusztai-Carey, M.; Russo, P.S.; Butko, P. A detergent-like mechanism of action of the cytolytic toxin Cyt1A from Bacillus thuringiensis var. israelensis. Biochemistry 2005, 44, 589–597. [Google Scholar] [CrossRef]
- Tharad, S.; Toca-Herrera, J.L.; Promdonkoy, B.; Krittanai, C. Bacillus thuringiensis Cyt2Aa2 toxin disrupts cell membranes by forming large protein aggregates. Biosci. Rep. 2016, 36. [Google Scholar] [CrossRef] [Green Version]
- Promdonkoy, B.; Rungrod, A.; Promdonkoy, P.; Pathaichindachote, W.; Krittanai, C.; Panyim, S. Amino acid substitutions in alphaA and alphaC of Cyt2Aa2 alter hemolytic activity and mosquito-larvicidal specificity. J. Biotechnol. 2008, 133, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Promdonkoy, B.; Pathaichindachote, W.; Krittanai, C.; Audtho, M.; Chewawiwat, N.; Panyim, S. Trp132, Trp154, and Trp157 are essential for folding and activity of a Cyt toxin from Bacillus thuringiensis. Biochem. Biophys. Res. Commun. 2004, 317, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Pathaichindachote, W.; Rungrod, A.; Audtho, M.; Soonsanga, S.; Krittanai, C.; Promdonkoy, B. Isoleucine at position 150 of Cyt2Aa toxin from Bacillus thuringiensis plays an important role during membrane binding and oligomerization. BMB Rep. 2013, 46, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Suktham, K.; Pathaichindachote, W.; Promdonkoy, B.; Krittanai, C. Essential role of amino acids in alphaD-beta4 loop of a Bacillus thuringiensis Cyt2Aa2 toxin in binding and complex formation on lipid membrane. Toxicon 2013, 74, 130–137. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Leidl, K.; Liebisch, G.; Richter, D.; Schmitz, G. Mass spectrometric analysis of lipid species of human circulating blood cells. Biochim. Biophys. Acta 2008, 1781, 655–664. [Google Scholar] [CrossRef]
- Stewart, S.E.; Bird, C.H.; Tabor, R.F.; D’Angelo, M.E.; Piantavigna, S.; Whisstock, J.C.; Trapani, J.A.; Martin, L.L.; Bird, P.I. Analysis of Perforin Assembly by Quartz Crystal Microbalance Reveals a Role for Cholesterol and Calcium-independent Membrane Binding. J. Biol. Chem. 2015, 290, 31101–31112. [Google Scholar] [CrossRef] [Green Version]
- Rabinovich, A.L.; Kornilov, V.V.; Balabaev, N.K.; Leermakers, F.A.M.; Filippov, A.V. Properties of unsaturated phospholipid bilayers: Effect of cholesterol. Biochem. (Moscow) Suppl. Ser. A Membr. Cell Biol. 2007, 1, 343–357. [Google Scholar] [CrossRef]
- Marheineke, K.; Grunewald, S.; Christie, W.; Reilander, H. Lipid composition of Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett. 1998, 441, 49–52. [Google Scholar] [CrossRef] [Green Version]
- Giocondi, M.-C.; Seantier, B.; Dosset, P.; Milhiet, P.-E.; Le Grimellec, C. Characterizing the interactions between GPI-anchored alkaline phosphatases and membrane domains by AFM. Pflügers Arch. -Eur. J. Physiol. 2008, 456, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Crespo-Villanueva, A.; Gumi-Audenis, B.; Sanz, F.; Artzner, F.; Meriadec, C.; Rousseau, F.; Lopez, C.; Giannotti, M.I.; Guyomarc’h, F. Casein interaction with lipid membranes: Are the phase state or charge density of the phospholipids affecting protein adsorption? Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 2588–2598. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Cai, M.; Xu, H.; Wang, H. Studying the membrane structure of chicken erythrocytes by in situ atomic force microscopy. Anal. Methods 2014, 6, 8115–8119. [Google Scholar] [CrossRef]
- Picas, L.; Rico, F.; Deforet, M.; Scheuring, S. Structural and mechanical heterogeneity of the erythrocyte membrane reveals hallmarks of membrane stability. ACS Nano 2013, 7, 1054–1063. [Google Scholar] [CrossRef] [PubMed]





| Lipid Composition (Mole Ratio) | ΔF5 (Hz) | ΔD5 (10−6) | Lipid Binding Rate, Γ (min) | |||
|---|---|---|---|---|---|---|
| WT | T144A | WT | T144A | WT | T144A | |
| POPC | −33.0 ± 3.8 | 1.3 ± 0.4 | 2.9 ± 1.1 | 0.0 ± 0.2 | 9.5 ± 0.3 | No binding |
| 1:0.4 POPC/Chol | −38.0 ± 1.7 | −29.1 ± 0.6 | 3.1 ± 1.0 | 2.3 ± 0.7 | 2.1 ± 0.2 | 11.2 ± 2.0 |
| 1:1 SM/POPC | −40.1 ± 3.4 | −24.4 ± 10.6 | 5.0 ± 0.2 | 3.0 ± 2.2 | 2.2 ± 0.6 | 52.1 ± 27.3 |
| 1:1:1 SM/POPC/Chol | −30.2 ± 4.4 | −25.4 ± 2.4 | 2.3 ± 2.0 | 2.2 ± 0.2 | 2.5 ± 0.4 | 10.9 ± 1.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tharad, S.; Promdonkoy, B.; Toca-Herrera, J.L. Protein-Lipid Interaction of Cytolytic Toxin Cyt2Aa2 on Model Lipid Bilayers of Erythrocyte Cell Membrane. Toxins 2020, 12, 226. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12040226
Tharad S, Promdonkoy B, Toca-Herrera JL. Protein-Lipid Interaction of Cytolytic Toxin Cyt2Aa2 on Model Lipid Bilayers of Erythrocyte Cell Membrane. Toxins. 2020; 12(4):226. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12040226
Chicago/Turabian StyleTharad, Sudarat, Boonhiang Promdonkoy, and José L. Toca-Herrera. 2020. "Protein-Lipid Interaction of Cytolytic Toxin Cyt2Aa2 on Model Lipid Bilayers of Erythrocyte Cell Membrane" Toxins 12, no. 4: 226. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12040226