Transcriptomics Reveals the Effect of Thymol on the Growth and Toxin Production of Fusarium graminearum
Abstract
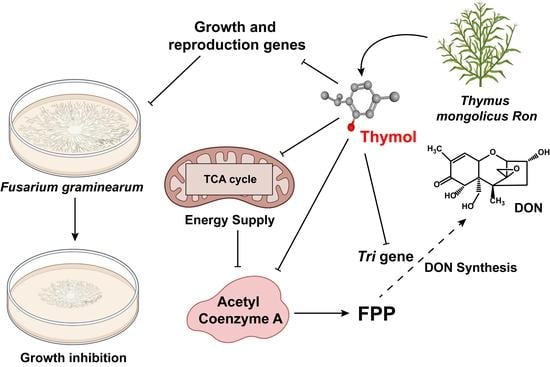
:1. Introduction
2. Results
2.1. The Effect of Thymol on the Growth of F. graminearum Hypha
2.2. The Effect of Thymol on DON Production by F. graminearum
2.3. The Effect of Thymol on the Transcriptome of F. graminearum
2.4. Unigenes Related to Mycelial Growth and DON Production
3. Discussion
3.1. The Effect of Thymol on the Growth of Mycelium
3.2. The Effect of Thymol on DON Production by F. graminearum
3.3. The Effect of Thymol on Glycolysis in F. graminearum
4. Conclusions
5. Materials and Methods
5.1. Fungal Strain, Media and Culture Condition
5.2. Determination of the Sensitivity of Mycelial Growth to Thymol
5.3. Changes in DON and 3-Ac-DON
5.4. Transcriptome Analysis
5.5. qRT-PCR
5.6. Statistical Analysis
5.7. Availability of Supporting Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board
Data Availability Statement
Conflicts of Interest
References
- Vandicke, J.; De Visschere, K.; Ameye, M.; Croubels, S.; De Saeger, S.; Audenaert, K.; Haesaert, G. Multi-Mycotoxin Contamination of Maize Silages in Flanders, Belgium: Monitoring Mycotoxin Levels from Seed to Feed. Toxins 2021, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Mielniczuk, E.; Skwaryło-Bednarz, B. Fusarium Head Blight, mycotoxins and strategies for their reduction. Agronomy 2020, 10, 509. [Google Scholar] [CrossRef] [Green Version]
- Bhat, R.; Rai, R.V.; Karim, A.A. Mycotoxins in Food and Feed: Present Status and Future Concerns. Compr. Rev. Food Sci. Food Saf. 2010, 9, 57–81. [Google Scholar] [CrossRef]
- Gruber-Dorninger, C.; Jenkins, T.; Schatzmayr, G. Global mycotoxin occurrence in feed: A ten-year survey. Toxins 2019, 11, 375. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Jia, S.; Zhang, J.; Zhang, C.; Wang, S.; Rajput, S.A.; Sun, L.; Qi, D. Transcriptomics and flow cytometry reveals the cytotoxicity of aflatoxin B1 and aflatoxin M1 in bovine mammary epithelial cells. Ecotoxicol. Environ. Saf. 2021, 209, 111823. [Google Scholar] [CrossRef]
- Wu, K.; Liu, M.; Wang, H.; Rajput, S.A.; Shan, Y.; Qi, D.; Wang, S. The Mechanism Underlying the Extreme Sensitivity of Duck to Aflatoxin B1. Oxid. Med. Cell. Longev. 2021, 2021, 9996503. [Google Scholar] [CrossRef]
- Wu, K.; Ren, C.; Gong, Y.; Gao, X.; Rajput, S.A.; Qi, D.; Wang, S. The insensitive mechanism of poultry to zearalenone: A review. Anim. Nutr. 2021, 7, 587–594. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Xue, D.; Zhang, C.; Rajput, S.A.; Qi, D. Mechanism of deoxynivalenol mediated gastrointestinal toxicity: Insights from mitochondrial dysfunction. Food Chem. Toxicol. 2021, 153, 112214. [Google Scholar] [CrossRef]
- Khoshal, A.K.; Novak, B.; Martin, P.; Jenkins, T.; Neves, M.; Schatzmayr, G.; Oswald, I.P.; Pinton, P. Co-occurrence of DON and Emerging Mycotoxins in Worldwide Finished Pig Feed and Their Combined Toxicity in Intestinal Cells. Toxins 2019, 11, 727. [Google Scholar] [CrossRef] [Green Version]
- Audenaert, K.; Callewaert, E.; Hofte, M.; De Saeger, S.; Haesaert, G. Hydrogen peroxide induced by the fungicide prothioconazole triggers deoxynivalenol (DON) production by Fusarium graminearum. BMC Microbiol. 2010, 10, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Z.; Wang, X.; Shen, Y.; Thakur, K.; Han, J.; Zhang, J.; Hu, F.; Wei, Z. Recent updates on the chemistry, bioactivities, mode of action, and industrial applications of plant essential oils. Trends Food Sci. Technol. 2021, 110, 78–89. [Google Scholar] [CrossRef]
- Prakash, B.; Kedia, A.; Mishra, P.K.; Dubey, N.K. Plant essential oils as food preservatives to control moulds, mycotoxin contamination and oxidative deterioration of agri-food commodities—Potentials and challenges. Food Control 2015, 47, 381–391. [Google Scholar] [CrossRef]
- Hu, L.B.; Ban, F.F.; Li, H.B.; Qian, P.P.; Shen, Q.S.; Zhao, Y.Y.; Mo, H.Z.; Zhou, X. Thymol Induces Conidial Apoptosis in Aspergillus flavus via Stimulating K(+) Eruption. J. Agric. Food Chem. 2018, 66, 8530–8536. [Google Scholar] [CrossRef]
- Wang, K.; Jiang, S.; Yang, Y.; Fan, L.; Su, F.; Ye, M. Synthesis and antifungal activity of carvacrol and thymol esters with heteroaromatic carboxylic acids. Nat. Prod. Res. 2019, 33, 1924–1930. [Google Scholar] [CrossRef]
- Kong, J.; Zhang, Y.; Ju, J.; Xie, Y.; Guo, Y.; Cheng, Y.; Qian, H.; Quek, S.Y.; Yao, W. Antifungal effects of thymol and salicylic acid on cell membrane and mitochondria of Rhizopus stolonifer and their application in postharvest preservation of tomatoes. Food Chem. 2019, 285, 380–388. [Google Scholar] [CrossRef]
- Kong, J.; Xie, Y.; Yu, H.; Guo, Y.; Cheng, Y.; Qian, H.; Yao, W. Synergistic antifungal mechanism of thymol and salicylic acid on Fusarium solani. LWT 2021, 140, 110787. [Google Scholar] [CrossRef]
- Perczak, A.; Gwiazdowska, D.; Marchwińska, K.; Juś, K.; Gwiazdowski, R.; Waśkiewicz, A. Antifungal activity of selected essential oils against Fusarium culmorum and F. graminearum and their secondary metabolites in wheat seeds. Arch. Microbiol. 2019, 201, 1085–1097. [Google Scholar] [CrossRef] [Green Version]
- Velluti, A.; Sanchis, V.; Ramos, A.J.; Turon, C.; Marin, S. Impact of essential oils on growth rate, zearalenone and deoxynivalenol production by Fusarium graminearum under different temperature and water activity conditions in maize grain. J. Appl. Microbiol. 2004, 96, 716–724. [Google Scholar] [CrossRef]
- Wei, J.T.; Wu, K.T.; Sun, H.; Khalil, M.M.; Dai, J.F.; Liu, Y.; Liu, Q.; Zhang, N.Y.; Qi, D.S.; Sun, L.H. A Novel Modified Hydrated Sodium Calcium Aluminosilicate (HSCAS) Adsorbent Can Effectively Reduce T-2 Toxin-Induced Toxicity in Growth Performance, Nutrient Digestibility, Serum Biochemistry, and Small Intestinal Morphology in Chicks. Toxins 2019, 11, 199. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Ge, J.; Yu, X. Transcriptome Analysis Reveals the Mechanism of Fungicidal of Thymol Against Fusarium oxysporum f. sp. niveum. Curr. Microbiol. 2018, 75, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Zhou, H.; Zhou, W.; Hu, L.; Chen, J.; Shi, Z. The Fungicidal Activity of Thymol against Fusarium graminearum via Inducing Lipid Peroxidation and Disrupting Ergosterol Biosynthesis. Molecules 2016, 21, 770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Ling, T.; Xue, Y.; Xu, C.; Zhou, W.; Hu, L.; Chen, J.; Shi, Z. Thymol Mitigates Cadmium Stress by Regulating Glutathione Levels and Reactive Oxygen Species Homeostasis in Tobacco Seedlings. Molecules 2016, 21, 1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, X.; Wang, Y.; Jiang, P.; Quek, S. Antibacterial activity and mechanism of cinnamon essential oil against Escherichia coli and Staphylococcus aureus. Food Control 2016, 59, 282–289. [Google Scholar] [CrossRef]
- Mith, H.; Dure, R.; Delcenserie, V.; Zhiri, A.; Daube, G.; Clinquart, A. Antimicrobial activities of commercial essential oils and their components against food-borne pathogens and food spoilage bacteria. Food Sci. Nutr. 2014, 2, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Hu, W.; Deng, J.; Liu, X.; Zhou, J.; Li, X. Antibacterial activity of Litsea cubeba essential oil and its mechanism against Botrytis cinerea. Rsc Adv. 2019, 9, 28987–28995. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.H.; Zhang, Z.H.; Zeng, X.A.; Gong, D.M.; Wang, M.S. Combination of microbiological, spectroscopic and molecular docking techniques to study the antibacterial mechanism of thymol against Staphylococcus aureus: Membrane damage and genomic DNA binding. Anal. Bioanal. Chem. 2017, 409, 1615–1625. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, Z.; Zhong, H.; Wang, S.; Yang, W.; Liu, Y.; Wang, S. RNA-Seq-based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins 2014, 6, 3187–3207. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.Q.; Zhao, X.X.; Zhi, Q.Q.; Zhao, M.; He, Z.M. Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 2013, 56, 78–86. [Google Scholar] [CrossRef]
- Kachur, K.; Suntres, Z. The antibacterial properties of phenolic isomers, carvacrol and thymol. Crit. Rev. Food Sci. Nutr. 2020, 60, 3042–3053. [Google Scholar] [CrossRef]
- Marchese, A.; Orhan, I.E.; Daglia, M.; Barbieri, R.; Di Lorenzo, A.; Nabavi, S.F.; Gortzi, O.; Izadi, M.; Nabavi, S.M. Antibacterial and antifungal activities of thymol: A brief review of the literature. Food Chem. 2016, 210, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.M.; Ribeiro, R.; Martins, M.L.; Antonio, D.S.C.T.; Santana, F.C.; Lirian, J.C.; Borges, W.S.; Costa, A.V.; Queiroz, V.T.; Scherer, R.; et al. Thymol as an Interesting Building Block for Promising Fungicides against Fusarium solani. J. Agric. Food Chem. 2021, 69, 6958–6967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, H.; Zhu, W.; Li, W.; Wang, F. Transcriptome Analysis Reveals the Effects of Chinese Chive (Allium tuberosum R.) Extract on Fusarium oxysporum f. sp. radicis-lycopersici Spore Germination. Curr. Microbiol. 2020, 77, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Oufensou, S.; Balmas, V.; Azara, E.; Fabbri, D.; Dettori, M.A.; Schuller, C.; Zehetbauer, F.; Strauss, J.; Delogu, G.; Migheli, Q. Naturally Occurring Phenols Modulate Vegetative Growth and Deoxynivalenol Biosynthesis in Fusarium graminearum. ACS Omega 2020, 5, 29407–29415. [Google Scholar] [CrossRef]
- Cui, W.; Du, K.; Ling, Y.; Yang, C. Activity of eugenol derivatives against Fusarium graminearum Q1 strain and screening of isoeugenol mixtures. J. Plant Pathol. 2021, 103, 915–921. [Google Scholar] [CrossRef]
- Venturini, T.P.; Rossato, L.; Chassot, F.; De Azevedo, M.I.; Al-Hatmi, A.M.S.; Santurio, J.M.; Alves, S.H. Activity of cinnamaldehyde, carvacrol and thymol combined with antifungal agents against Fusarium spp. J. Essent. Oil Res. 2021, 33, 502–508. [Google Scholar] [CrossRef]
- Khmelnitskiy, G.O.; Korzunenko, V.D. Effect of combined sorbent preparation on growth, performance and haematobiochemical alterations in broiler chickens under the influence of T-2 toxin and deoxynivalenol mixed toxicosis. Bangladesh J. Vet. Med. 2013, 11, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [Green Version]
- Adnan, M.; Fang, W.; Sun, P.; Zheng, Y.; Abubakar, Y.S.; Zhang, J.; Lou, Y.; Zheng, W.; Lu, G.D. R-SNARE FgSec22 is essential for growth, pathogenicity and DON production of Fusarium graminearum. Curr. Genet. 2020, 66, 421–435. [Google Scholar] [CrossRef]
- Shin, J.; Kim, J.E.; Lee, Y.W.; Son, H. Fungal Cytochrome P450s and the P450 Complement (CYPome) of Fusarium graminearum. Toxins 2018, 10, 112. [Google Scholar] [CrossRef] [Green Version]
- Bombassaro, A.; Schneider, G.X.; Costa, F.F.; Leao, A.; Soley, B.S.; Medeiros, F.; Da, S.N.; Lima, B.; Castro, R.; Bocca, A.L.; et al. Genomics and Virulence of Fonsecaea pugnacius, Agent of Disseminated Chromoblastomycosis. Front. Genet. 2020, 11, 822. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; McCormick, S.; Tiley, H.; Usgaard, T. Detoxification and Excretion of Trichothecenes in Transgenic Arabidopsisthaliana Expressing Fusarium graminearum Trichothecene 3-O-acetyltransferase. Toxins 2021, 13, 320. [Google Scholar] [CrossRef] [PubMed]
- Oufensou, S.; Dessì, A.; Dallocchio, R.; Balmas, V.; Azara, E.; Carta, P.; Migheli, Q.; Delogu, G. Molecular Docking and Comparative Inhibitory Efficacy of Naturally Occurring Compounds on Vegetative Growth and Deoxynivalenol Biosynthesis in Fusarium culmorum. Toxins 2021, 13, 759. [Google Scholar] [CrossRef] [PubMed]
- Miladi, H.; Zmantar, T.; Chaabouni, Y.; Fedhila, K.; Bakhrouf, A.; Mahdouani, K.; Chaieb, K. Antibacterial and efflux pump inhibitors of thymol and carvacrol against food-borne pathogens. Microb. Pathog. 2016, 99, 95–100. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Yang, N.; Wen, Z.; Sun, X.; Chai, Y.; Ma, Z. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun. 2018, 9, 3429. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; van Diepeningen, A.D.; van der Lee, T.; Waalwijk, C.; Xu, J.; Xu, J.; Zhang, H.; Chen, W.; Feng, J. The Fusarium graminearum Histone Acetyltransferases Are Important for Morphogenesis, DON Biosynthesis, and Pathogenicity. Front. Microbiol. 2018, 9, 654. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Min, K.; Son, H.; Park, A.R.; Kim, J.C.; Choi, G.J.; Lee, Y.W. ELP3 is involved in sexual and asexual development, virulence, and the oxidative stress response in Fusarium graminearum. Mol. Plant Microbe Interact. 2014, 27, 1344–1355. [Google Scholar] [CrossRef] [Green Version]
- Wen, N.; Liu, W.; Hou, Y.; Zhao, Z. The kinetics behavior of the reduction of formaldehyde catalyzed by Alcohol Dehydrogenase (ADH) and partial uncompetitive substrate inhibition by NADH. Appl. Biochem. Biotechnol. 2013, 170, 370–380. [Google Scholar] [CrossRef]
- Fujita, H.; Aoki, H.; Ajioka, I.; Yamazaki, M.; Abe, M.; Oh-Nishi, A.; Sakimura, K.; Sugihara, I. Detailed expression pattern of aldolase C (Aldoc) in the cerebellum, retina and other areas of the CNS studied in Aldoc-Venus knock-in mice. PLoS ONE 2014, 9, e86679. [Google Scholar] [CrossRef]
- Li, Y.; Ma, C.; Li, W.; Yang, Y.; Li, X.; Liu, J.; Wang, J.; Li, S.; Liu, Y.; Li, K.; et al. A missense variant in NDUFA6 confers schizophrenia risk by affecting YY1 binding and NAGA expression. Mol. Psychiatry 2021. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, J.; Shen, F.; Ma, X.; Wang, Z.; Hou, X.; Hao, E.; Hou, Y.; Bai, G. Cinnamaldehyde changes the dynamic balance of glucose metabolism by targeting ENO1. Life Sci. 2020, 258, 118151. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Bai, M.; Sun, Y.; Abdel-Samie, M.A.; Lin, L. Antibacterial activity and mechanism of Chuzhou chrysanthemum essential oil. J. Funct. Foods 2018, 48, 159–166. [Google Scholar] [CrossRef]
- Tian, J.; Ban, X.; Zeng, H.; He, J.; Huang, B.; Wang, Y. Chemical composition and antifungal activity of essential oil from Cicuta virosa L. var. latisecta Celak. Int. J. Food Microbiol. 2011, 145, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Lee, Y.; Oh, S.M.; Yoon, D.; Choi, D.J.; Kwon, D.; Kang, O.; Lee, D.Y. Inhibitory Effects of Thymol Isolated from Curcuma longa L. on Adipogenesis in HepG2 Cells. Processes 2020, 8, 1191. [Google Scholar] [CrossRef]
- Araniti, F.; Miras-Moreno, B.; Lucini, L.; Landi, M.; Abenavoli, M.R. Metabolomic, proteomic and physiological insights into the potential mode of action of thymol, a phytotoxic natural monoterpenoid phenol. Plant Physiol. Biochem. 2020, 153, 141–153. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Z.; Mo, H.; Zhao, Y.; Li, H.; Zhang, H.; Hu, L.; Zhou, X. Thymol Mediates Bactericidal Activity against Staphylococcus aureus by Targeting an Aldo–Keto Reductase and Consequent Depletion of NADPH. J. Agric. Food Chem. 2019, 67, 8382–8392. [Google Scholar] [CrossRef]
- Ban, F.; Hu, L.; Zhou, X.H.; Zhao, Y.; Mo, H.; Li, H.; Zhou, W. Inverse molecular docking reveals a novel function of thymol: Inhibition of fat deposition induced by high-dose glucose in Caenorhabditis elegans. Food Sci. Nutr. 2021, 9, 4243–4253. [Google Scholar] [CrossRef]
- Baldissera, M.D.; Souza, C.F.; De Matos, A.; Baldisserotto, B.; Da, S.A.; Monteiro, S.G. Tissue oxidative damage mediates impairment on phosphotransfer network during thymol intake: Effects on hepatic and renal bioenergetics. Chem. Biol. Interact. 2018, 296, 83–88. [Google Scholar] [CrossRef]
- Wang, S.; Yang, J.; Zhang, B.; Wu, K.; Yang, A.; Li, C.; Zhang, J.; Zhang, C.; Rajput, S.A.; Zhang, N.; et al. Deoxynivalenol Impairs Porcine Intestinal Host Defense Peptide Expression in Weaned Piglets and IPEC-J2 Cells. Toxins 2018, 10, 541. [Google Scholar] [CrossRef] [Green Version]
- Hellin, P.; King, R.; Urban, M.; Hammond-Kosack, K.E.; Legreve, A. The adaptation of Fusarium culmorum to DMI Fungicides Is Mediated by Major Transcriptome Modifications in Response to Azole Fungicide, Including the Overexpression of a PDR Transporter (FcABC1). Front. Microbiol. 2018, 9, 1385. [Google Scholar] [CrossRef]
- Liu, X.; Yu, F.; Schnabel, G.; Wu, J.; Wang, Z.; Ma, Z. Paralogous cyp51 genes in Fusarium graminearum mediate differential sensitivity to sterol demethylation inhibitors. Fungal Genet. Biol. 2011, 48, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Duan, Y.; Bian, C.; Pan, X.; Yao, C.; Wang, J.; Zhou, M. Effects of validamycin in controlling Fusarium head blight caused by Fusarium graminearum: Inhibition of DON biosynthesis and induction of host resistance. Pestic. Biochem. Physiol. 2019, 153, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Stroka, J.; Derbyshire, M.; Mischke, C.; Ambrosio, M.; Kroeger, K.; Arranz, I.; Sizoo, E.; van Egmond, H. Liquid chromatographic determination of deoxynivalenol in baby food and animal feed: Interlaboratory study. J. AOAC Int. 2006, 89, 1012–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, X.; Han, Y.; Liu, C. The Fungicidal Activity of Tebuconazole Enantiomers against Fusarium graminearum and Its Selective Effect on DON Production under Different Conditions. J. Agric. Food Chem. 2018, 66, 3637–3643. [Google Scholar] [CrossRef]





| Name | COG_ID | FC | Swiss-Prot_Description | Functional Categories |
|---|---|---|---|---|
| CND2 | COG5229 | 2.37 | Condensin complex subunit 2 | Chromatin structure and dynamics |
| MPIP | COG5105 | 3.05 | M-phase inducer phosphatase | Cell cycle control, cell division, chromosome partitioning |
| HSK1 | ENOG410YKCM | 3.25 | Cell cycle serine/threonine-protein kinase hsk1 | |
| CDC45 | ENOG410XR8E | 2.00 | Cell division control protein 45 homolog | |
| APC10 | COG5156 | 3.50 | Anaphase-promoting complex subunit 10 | |
| CALM | COG5126 | 2.98 | Calmodulin | |
| HXT2 | ENOG410XNQK | 0.46 | High-affinity glucose transporter HXT2 | Carbohydrate transport and metabolism |
| PHSG | COG0058 | 2.41 | Glycogen phosphorylase | |
| HSP80 | COG0326 | 0.06 | Heat shock cognate protein 80 | Coenzyme transport and metabolism |
| FSR4 | ENOG4111F9D | 2.44 | Trans-enoyl reductase fsr4 | Lipid transport and metabolism |
| PARP2 | ENOG410XP18 | 13.54 | ADP-ribose polymerase 2 | Translation, ribosomal structure, and biogenesis |
| SMC2 | COG1196 | 2.79 | Structural maintenance of chromosomes protein 2 | Transcription |
| SAK1 | ENOG410XSHE | 2.00 | Protein sak1 | |
| MCM1 | COG5068 | 2.02 | Transcription factor of morphogenesis MCM1 | |
| ORC4 | ENOG410XSK0 | 2.70 | Origin recognition complex subunit 4 | Replication, recombination, and repair |
| RAD1 | ENOG410YHQU | 2.26 | DNA damage checkpoint control protein rad1 | |
| SKP1 | COG5201 | 2.42 | E3 ubiquitin ligase complex SCF subunit scon-3 | Posttranslational modification, protein turnover |
| CDK1 | ENOG410XPP3 | 2.26 | Cyclin-dependent kinase 1 | Signal transduction mechanisms |
| PPZ | COG0639 | 0.49 | Serine/threonine-protein phosphatase PP-Z | |
| PHLN | COG3511 | 0.08 | Non-hemolytic phospholipase C | Cell wall/membrane/envelope biogenesis |
| GAL10 | COG1087 | 3.03 | Bifunctional protein GAL10 |
| Name | COG_ID | FC | Swiss-Prot_Description | Functional Categories |
|---|---|---|---|---|
| TRI4 | COG2124 | 0.01 | Cytochrome P450 monooxygenase | Cytochrome P450 |
| TRI13 | COG2124 | 0.02 | Cytochrome P450 monooxygenase | |
| TRI11 | COG2124 | 0.02 | Trichothecene C-15 hydroxylase | |
| TRI1 | COG2124 | 0.01 | Cytochrome P450 monooxygenase | |
| TRI8 | ENOG410YF7H | 0.01 | Trichothecene C-3 esterase | Trichothecene C-3 esterase |
| TRI6 | ENOG410YRUA | 0.01 | Trichothecene biosynthesis transcription regulator 6 | Trichothecene biosynthesis transcription regulator |
| TRI5 | ENOG410YBAU | 0.01 | Trichodiene synthase | Trichodiene synthase |
| CLM1 | ENOG410YEIS | 0.01 | Longiborneol synthase CLM1 | |
| TRI3 | ENOG4112AIP | 0.01 | Trichothecene 15-O-acetyltransferase | Trichothecene 15-O-acetyltransferase |
| TRI14 | ENOG41118DK | 0.03 | Core trichothecene cluster | Core trichothecene cluster |
| TR101 | ENOG410YJA8 | 0.09 | Trichothecene 3-O-acetyltransferase | Transferase family |
| TRI10 | ENOG410YFZP | 0.06 | Trichothecene biosynthesis transcription regulator 10 | Fungal trichothecene efflux pump |
| TRI12 | ENOG410XNQK | 0.01 | Trichothecene efflux pump TRI12 | |
| ACLA | ENOG410XNQK | 0.01 | MFS efflux transporter aclA | |
| STR3 | ENOG41109Q9 | 0.06 | Siderophore iron transporter 3 | |
| LACP | ENOG410XNQK | 0.08 | Lactose permease | |
| NAG3 | COG0477 | 0.00 | Major facilitator superfamily multidrug transporter NAG3 | |
| GSFJ | ENOG410XNQK | 0.08 | Probable efflux pump gsfJ | |
| YJ94 | ENOG410ZNSM | 0.00 | Uncharacterized membrane protein | |
| MF227 | ENOG4111EZJ | 0.02 | Probable efflux pump mfs2 | |
| ATB_A | COG0477 | 0.07 | Efflux pump atB | |
| FUS6 | ENOG410XNQK | 0.06 | Efflux pump FUS6 | |
| FUB11 | COG0477 | 0.06 | Efflux pump FUBT | |
| MIRB | ENOG410XNQK | 0.01 | Siderophore iron transporter mirB | |
| ROQT | ENOG410XNQK | 0.03 | Efflux pump roqT | |
| SIT1 | ENOG410ZYHG | 0.15 | Siderophore iron transporter 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.-Q.; Wu, K.-T.; Yang, P.; Hou, F.; Rajput, S.A.; Qi, D.-S.; Wang, S. Transcriptomics Reveals the Effect of Thymol on the Growth and Toxin Production of Fusarium graminearum. Toxins 2022, 14, 142. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins14020142
Wang L-Q, Wu K-T, Yang P, Hou F, Rajput SA, Qi D-S, Wang S. Transcriptomics Reveals the Effect of Thymol on the Growth and Toxin Production of Fusarium graminearum. Toxins. 2022; 14(2):142. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins14020142
Chicago/Turabian StyleWang, Lian-Qun, Kun-Tan Wu, Ping Yang, Fang Hou, Shahid Ali Rajput, De-Sheng Qi, and Shuai Wang. 2022. "Transcriptomics Reveals the Effect of Thymol on the Growth and Toxin Production of Fusarium graminearum" Toxins 14, no. 2: 142. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins14020142