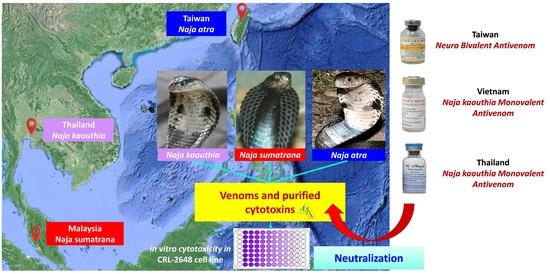
Cytotoxicity of Venoms and Cytotoxins from Asiatic Cobras (Naja kaouthia, Naja sumatrana, Naja atra) and Neutralization by Antivenoms from Thailand, Vietnam, and Taiwan
Abstract
:1. Introduction
2. Results
2.1. Isolation and Validation of Cobra Cytotoxins
2.2. Cytotoxicity of Cobra Venoms and Purified Cytotoxins
2.3. Neutralization of Cytotoxicity by Antivenoms
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Venoms and Antivenoms
5.2. Resource S Cation-Exchange and C18 Reverse-Phase High-Performance Liquid Chromatography
5.3. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
5.4. Nano-Electrospray Ionization Liquid Chromatography-Tandem Mass Spectrometry (Nano-ESI LC-MS/MS)
5.5. Multiple Sequence Alignment
5.6. Cell Culture
5.7. In Vitro Cytotoxicity Induced by Cobra Venoms and CTX
5.8. Neutralization Assay of Cytotoxicity
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gutierrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17079. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global snakebite burden. In Proceedings of the Seventy-First World Health Assembly, Geneva, Switzerland, 21–26 May 2018. [Google Scholar]
- WHO. Guidelines for the Management of Snake Bites; Regional office for South-East Asia: New Delhi, India, 2016. [Google Scholar]
- WHO. Snakebite Envenoming: A Strategy for Prevention and Control; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Tan, N.H.; Tan, K.Y.; Tan, C.H. Snakebite in Southeast Asia: Envenomation and Clinical Management. In Handbook of Venoms and Toxins of Reptiles, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Wong, O.F.; Lam, T.S.; Fung, H.T.; Choy, C.H. Five-year experience with Chinese cobra (Naja atra)—Related injuries in two acute hospitals in Hong Kong. Hong Kong Med. J. 2010, 16, 36–43. [Google Scholar] [PubMed]
- Ang, L.J.; Sanjay, S.; Sangtam, T. Ophthalmia due to spitting cobra venom in an urban setting--a report of three cases. Middle East Afr. J. Ophthalmol. 2014, 21, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Chu, E.R.; Weinstein, S.A.; White, J.; Warrell, D.A. Venom ophthalmia caused by venoms of spitting elapid and other snakes: Report of ten cases with review of epidemiology, clinical features, pathophysiology and management. Toxicon 2010, 56, 259–272. [Google Scholar] [CrossRef]
- Tsai, T.H.; Lin, C.C.; Mao, Y.C.; Hung, C.L.; Yang, Y.C.; Yang, C.C.; Jeng, M.J. Naja atra venom-spit ophthalmia in Taiwan: An epidemiological survey from 1990 to 2016. J. Chin. Med. Assoc. 2020, 83, 77–83. [Google Scholar] [CrossRef]
- Tan, K.Y.; Wong, K.Y.; Tan, N.H.; Tan, C.H. Quantitative proteomics of Naja annulifera (sub-Saharan snouted cobra) venom and neutralization activities of two antivenoms in Africa. Int. J. Biol. Macromol. 2020, 158, 605–616. [Google Scholar] [CrossRef]
- Wong, K.Y.; Tan, K.Y.; Tan, N.H.; Tan, C.H. A Neurotoxic Snake Venom without Phospholipase A2: Proteomics and Cross-Neutralization of the Venom from Senegalese Cobra, Naja senegalensis (Subgenus: Uraeus). Toxins 2021, 13, 60. [Google Scholar] [CrossRef]
- Huang, H.W.; Liu, B.S.; Chien, K.Y.; Chiang, L.C.; Huang, S.Y.; Sung, W.C.; Wu, W.G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteomics 2015, 128, 92–104. [Google Scholar] [CrossRef]
- Antil-Delbeke, S.; Gaillard, C.; Tamiya, T.; Corringer, P.J.; Changeux, J.P.; Servent, D.; Menez, A. Molecular determinants by which a long chain toxin from snake venom interacts with the neuronal alpha 7-nicotinic acetylcholine receptor. J. Biol. Chem. 2000, 275, 29594–29601. [Google Scholar] [CrossRef] [Green Version]
- Kini, R.M. Molecular moulds with multiple missions: Functional sites in three-finger toxins. Clin. Exp. Pharmacol. Physiol. 2002, 29, 815–822. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Lesovoy, D.M.; Dubinnyi, M.A.; Konshina, A.G.; Utkin, Y.N.; Efremov, R.G.; Arseniev, A.S. Interaction of three-finger toxins with phospholipid membranes: Comparison of S- and P-type cytotoxins. Biochem. J. 2005, 387, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Konshina, A.G.; Boldyrev, I.A.; Utkin, Y.N.; Omel’kov, A.V.; Efremov, R.G. Snake Cytotoxins Bind to Membranes via Interactions with Phosphatidylserine Head Groups of Lipids. PLoS ONE 2011, 6, e19064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, K.Y.; Chiang, C.M.; Hseu, Y.C.; Vyas, A.A.; Rule, G.S.; Wu, W. Two distinct types of cardiotoxin as revealed by the structure and activity relationship of their interaction with zwitterionic phospholipid dispersions. J. Biol. Chem. 1994, 269, 14473–14483. [Google Scholar] [CrossRef]
- Chong, H.P.; Tan, K.Y.; Tan, C.H. Cytotoxicity of Snake Venoms and Cytotoxins From Two Southeast Asian Cobras (Naja sumatrana, Naja kaouthia): Exploration of Anticancer Potential, Selectivity, and Cell Death Mechanism. Front. Mol. Biosci. 2020, 7, 583587. [Google Scholar] [CrossRef] [PubMed]
- Attarde, S.S.; Pandit, S.V. Cytotoxic activity of NN-32 toxin from Indian spectacled cobra venom on human breast cancer cell lines. BMC Complement. Altern. Med. 2017, 17, 503. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ming, W.; Wang, Y.; Liu, S.; Qiu, Y.; Xiang, Y.; Hu, L.; Fan, L.; Peng, X.; Wang, H.; et al. Cytotoxin 1 from Naja atra Cantor venom induced necroptosis of leukemia cells. Toxicon 2019, 165, 110–115. [Google Scholar] [CrossRef]
- Liu, C.C.; Chou, Y.S.; Chen, C.Y.; Liu, K.L.; Huang, G.J.; Yu, J.S.; Wu, C.J.; Liaw, G.W.; Hsieh, C.H.; Chen, C.K. Pathogenesis of local necrosis induced by Naja atra venom: Assessment of the neutralization ability of Taiwanese freeze-dried neurotoxic antivenom in animal models. PLoS Negl. Trop. Dis. 2020, 14, e0008054. [Google Scholar] [CrossRef] [Green Version]
- Rivel, M.; Solano, D.; Herrera, M.; Vargas, M.; Villalta, M.; Segura, A.; Arias, A.S.; Leon, G.; Gutierrez, J.M. Pathogenesis of dermonecrosis induced by venom of the spitting cobra, Naja nigricollis: An experimental study in mice. Toxicon 2016, 119, 171–179. [Google Scholar] [CrossRef]
- Hsieh, Y.H.; Hsueh, J.H.; Liu, W.C.; Yang, K.C.; Hsu, K.C.; Lin, C.T.; Ho, Y.Y.; Chen, L.W. Contributing Factors for Complications and Outcomes in Patients With Snakebite: Experience in a Medical Center in Southern Taiwan. Ann. Plast. Surg. 2017, 78, S32–S36. [Google Scholar] [CrossRef]
- Huang, L.-W.; Wang, J.-D.; Huang, J.-A.; Hu, S.-Y.; Wang, L.-M.; Tsan, Y.-T. Wound infections secondary to snakebite in central Taiwan. J. Venom. Anim. Toxins Incl. Trop. Dis. 2012, 18, 272–276. [Google Scholar] [CrossRef] [Green Version]
- WHO. Guidelines for the Management of Snake-Bites; World Health Organization: New Delhi, India, 2010. [Google Scholar]
- Gutiérrez, J.; Solano, G.; Pla, D.; Herrera, M.; Segura, Á.; Vargas, M.; Villalta, M.; Sánchez, A.; Sanz, L.; Lomonte, B.; et al. Preclinical Evaluation of the Efficacy of Antivenoms for Snakebite Envenoming: State-of-the-Art and Challenges Ahead. Toxins 2017, 9, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez, N.; Escalante, T.; Gutierrez, J.M.; Rucavado, A. Skin pathology induced by snake venom metalloproteinase: Acute damage, revascularization, and re-epithelization in a mouse ear model. J. Investig. Dermatol. 2008, 128, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Rudresha, G.V.; Urs, A.P.; Manjuprasanna, V.N.; Milan Gowda, M.D.; Jayachandra, K.; Rajaiah, R.; Vishwanath, B.S. Echis carinatus snake venom metalloprotease-induced toxicities in mice: Therapeutic intervention by a repurposed drug, Tetraethyl thiuram disulfide (Disulfiram). PLoS Negl. Trop. Dis. 2021, 15, e0008596. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Gunawardena, P.; Weilgama, D.; Maduwage, K.; Gawarammana, I. Comparative in-vivo toxicity of venoms from South Asian hump-nosed pit vipers (Viperidae: Crotalinae: Hypnale). BMC Res. Notes 2012, 5, 471. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, J.M.; Vargas, M.; Segura, A.; Herrera, M.; Villalta, M.; Solano, G.; Sanchez, A.; Herrera, C.; Leon, G. In vitro Tests for Assessing the Neutralizing Ability of Snake Antivenoms: Toward the 3Rs Principles. Front. Immunol. 2020, 11, 617429. [Google Scholar] [CrossRef]
- Kalam, Y.; Isbister, G.K.; Mirtschin, P.; Hodgson, W.C.; Konstantakopoulos, N. Validation of a cell-based assay to differentiate between the cytotoxic effects of elapid snake venoms. J. Pharmacol. Toxicol. Methods 2011, 63, 137–142. [Google Scholar] [CrossRef]
- Liu, B.S.; Wu, W.G.; Lin, M.H.; Li, C.H.; Jiang, B.R.; Wu, S.C.; Leng, C.H.; Sung, W.C. Identification of Immunoreactive Peptides of Toxins to Simultaneously Assess the Neutralization Potency of Antivenoms against Neurotoxicity and Cytotoxicity of Naja atra Venom. Toxins 2017, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Mendez, I.; Gutierrez, J.M.; Angulo, Y.; Calvete, J.J.; Lomonte, B. Comparative study of the cytolytic activity of snake venoms from African spitting cobras (Naja spp., Elapidae) and its neutralization by a polyspecific antivenom. Toxicon 2011, 58, 558–564. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Chanhome, L.; Tan, N.H. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: Elucidating geographical venom variation and insights into sequence novelty. PeerJ 2017, 5, e3142. [Google Scholar] [CrossRef] [Green Version]
- Chong, H.P.; Tan, K.Y.; Tan, N.H.; Tan, C.H. Exploring the Diversity and Novelty of Toxin Genes in Naja sumatrana, the Equatorial Spitting Cobra from Malaysia through De Novo Venom-Gland Transcriptomics. Toxins 2019, 11, 104. [Google Scholar] [CrossRef] [Green Version]
- Jayawardana, S.; Arambepola, C.; Chang, T.; Gnanathasan, A. Long-term health complications following snake envenoming. J. Multidiscip. Healthc. 2018, 11, 279–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranawaka, U.K.; Lalloo, D.G.; de Silva, H.J. Neurotoxicity in snakebite—The limits of our knowledge. PLoS Negl. Trop. Dis. 2013, 7, e2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.H.; Wong, K.Y.; Chong, H.P.; Tan, N.H.; Tan, K.Y. Proteomic insights into short neurotoxin-driven, highly neurotoxic venom of Philippine cobra (Naja philippinensis) and toxicity correlation of cobra envenomation in Asia. J. Proteomics 2019, 206, 103418. [Google Scholar] [CrossRef] [PubMed]
- Panagides, N.; Jackson, T.N.; Ikonomopoulou, M.P.; Arbuckle, K.; Pretzler, R.; Yang, D.C.; Ali, S.A.; Koludarov, I.; Dobson, J.; Sanker, B.; et al. How the Cobra Got Its Flesh-Eating Venom: Cytotoxicity as a Defensive Innovation and Its Co-Evolution with Hooding, Aposematic Marking, and Spitting. Toxins 2017, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Yap, M.K.; Fung, S.Y.; Tan, K.Y.; Tan, N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra). Acta Trop. 2014, 133, 15–25. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J. Proteomics 2015, 120, 105–125. [Google Scholar] [CrossRef]
- Petras, D.; Sanz, L.; Segura, A.; Herrera, M.; Villalta, M.; Solano, D.; Vargas, M.; Leon, G.; Warrell, D.A.; Theakston, R.D.; et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the pan-African EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J. Proteome Res. 2011, 10, 1266–1280. [Google Scholar] [CrossRef]
- WHO. Guidelines for the Prevention and Clinical Management of Snakebite in Africa; World Health Organization: Brazzaville, Congo, 2010. [Google Scholar]
- Blaylock, R.S.; Lichtman, A.R.; Potgieter, P.D. Clinical manifestations of Cape cobra (Naja nivea) bites. A report of 2 cases. S. Afr. Med. J. 1985, 68, 342–344. [Google Scholar]
- Botes, D.P.; Viljoen, C.C. The amino acid sequence of three non-curarimimetic toxins from Naja nivea venom. Biochim. Biophys. Acta (BBA) Protein Struct. 1976, 446, 1–9. [Google Scholar] [CrossRef]
- Joubert, F.J.; Taljaard, N. Naja haje haje (Egyptian cobra) venom. Some properties and the complete primary structure of three toxins (CM-2, CM-11 and CM-12). Eur. J. Biochem. 1978, 90, 359–367. [Google Scholar] [CrossRef]
- Weise, K.H.; Carlsson, F.H.; Joubert, F.J.; Strydom, D.J. Snake venom toxins. The purification of toxins VII1 and VII2, two cytotoxin homologues from banded Egyptian cobra (Naja haje annulifera) venom, and the complete amino acid sequence of toxin VII1. Hoppe Seylers Z. Physiol Chem 1973, 354, 1317–1326. [Google Scholar] [CrossRef]
- Tan, C.H.; Fung, S.Y.; Yap, M.K.; Leong, P.K.; Liew, J.L.; Tan, N.H. Unveiling the elusive and exotic: Venomics of the Malayan blue coral snake (Calliophis bivirgata flaviceps). J. Proteomics 2016, 132, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.M.; Chien, K.Y.; Lin, H.J.; Lin, J.F.; Yeh, H.C.; Ho, P.L.; Wu, W.G. Conformational change and inactivation of membrane phospholipid-related activity of cardiotoxin V from Taiwan cobra venom at acidic pH. Biochemistry 1996, 35, 9167–9176. [Google Scholar] [CrossRef]
- Watt, G.; Padre, L.; Tuazon, L.; Theakston, R.D.; Laughlin, L. Bites by the Philippine cobra (Naja naja philippinensis): Prominent neurotoxicity with minimal local signs. Am. J. Trop. Med. Hyg. 1988, 39, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.C.; Liu, P.Y.; Chiang, L.C.; Lai, C.S.; Lai, K.L.; Ho, C.H.; Wang, T.H.; Yang, C.C. Naja atra snakebite in Taiwan. Clin. Toxicol. 2018, 56, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Wongtongkam, N.; Wilde, H.; Sitthi-Amorn, C.; Ratanabanangkoon, K. A study of Thai cobra (Naja kaouthia) bites in Thailand. Mil. Med. 2005, 170, 336–341. [Google Scholar] [CrossRef] [Green Version]
- Paterna, A. Spitting behaviour in the Chinese cobra Naja atra. Herpetol. Bull. 2019, 22–25. [Google Scholar] [CrossRef]
- Wuster, W.; Thorpe, R.S. Dentitional Phenomena in Cobras Revisited: Spitting and Fang Structure in the Asiatic Species of Naja (Serpentes: Elapidae). Herpetologica 1992, 48, 424–434. [Google Scholar]
- Leong, P.K.; Fung, S.Y.; Tan, C.H.; Sim, S.M.; Tan, N.H. Immunological cross-reactivity and neutralization of the principal toxins of Naja sumatrana and related cobra venoms by a Thai polyvalent antivenom (Neuro Polyvalent Snake Antivenom). Acta Trop. 2015, 149, 86–93. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Neutralization of the Principal Toxins from the Venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into Toxin-Specific Neutralization by Two Different Antivenoms. Toxins 2016, 8, 86. [Google Scholar] [CrossRef] [Green Version]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Oh, A.M.F.; Tan, K.Y.; Tan, N.H.; Tan, C.H. Proteomics and neutralization of Bungarus multicinctus (Many-banded Krait) venom: Intra-specific comparisons between specimens from China and Taiwan. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021, 247, 109063. [Google Scholar] [CrossRef] [PubMed]
- Gasanov, S.E.; Dagda, R.K.; Rael, E.D. Snake Venom Cytotoxins, Phospholipase A2s, and Zn(2+)-dependent Metalloproteinases: Mechanisms of Action and Pharmacological Relevance. J. Clin. Toxicol. 2014, 4, 1000181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, K.Y.; Tan, C.H.; Tan, N.H. Venom and Purified Toxins of the Spectacled Cobra (Naja naja) from Pakistan: Insights into Toxicity and Antivenom Neutralization. Am. J. Trop. Med. Hyg. 2016, 94, 1392–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.H.; Wong, K.Y.; Tan, N.H.; Ng, T.S.; Tan, K.Y. Distinctive Distribution of Secretory Phospholipases A(2) in the Venoms of Afro-Asian Cobras (Subgenus: Naja, Afronaja, Boulengerina and Uraeus). Toxins 2019, 11, 116. [Google Scholar] [CrossRef] [Green Version]
- Ministry of Health Malaysia. Guideline: Management of Snakebite; Ministry of Health Malaysia: Putrajaya, Malaysia, 2017.
- Ratanabanangkoon, K.; Tan, K.Y.; Pruksaphon, K.; Klinpayom, C.; Gutierrez, J.M.; Quraishi, N.H.; Tan, C.H. A pan-specific antiserum produced by a novel immunization strategy shows a high spectrum of neutralization against neurotoxic snake venoms. Sci. Rep. 2020, 10, 11261. [Google Scholar] [CrossRef]
- Laustsen, A.H.; Johansen, K.H.; Engmark, M.; Andersen, M.R. Recombinant snakebite antivenoms: A cost-competitive solution to a neglected tropical disease? PLoS Negl. Trop. Dis. 2017, 11, e0005361. [Google Scholar] [CrossRef] [Green Version]
- Albulescu, L.O.; Xie, C.; Ainsworth, S.; Alsolaiss, J.; Crittenden, E.; Dawson, C.A.; Softley, R.; Bartlett, K.E.; Harrison, R.A.; Kool, J.; et al. A therapeutic combination of two small molecule toxin inhibitors provides broad preclinical efficacy against viper snakebite. Nat. Commun. 2020, 11, 6094. [Google Scholar] [CrossRef]
- Tan, C.H.; Lingam, T.M.C.; Tan, K.Y. Varespladib (LY315920) rescued mice from fatal neurotoxicity caused by venoms of five major Asiatic kraits (Bungarus spp.) in an experimental envenoming and rescue model. Acta Trop. 2021, 227, 106289. [Google Scholar] [CrossRef]
- Lazar, I.J.; Lazar, I.S. Gel Analyzer 2010a: Freeware 1D gel Electrophoresis Image Analysis Software. Available online: http://www.gelanalyzer.com (accessed on 11 May 2020).
- Tan, C.H.; Wong, K.Y.; Tan, K.Y.; Tan, N.H. Venom proteome of the yellow-lipped sea krait, Laticauda colubrina from Bali: Insights into subvenomic diversity, venom antigenicity and cross-neutralization by antivenom. J. Proteomics 2017, 166, 48–58. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omran, M.A.A.; Fabb, S.A.; Dickson, G. Biochemical and morphological analysis of cell death induced by Egyptian cobra (Naja haje) venom on cultured cells. J. Venom. Anim. Toxins Incl. Trop. Dis. 2004, 10, 219–241. [Google Scholar] [CrossRef]







| Protein Designated Identity | Protein Score | Protein Name Matched | Accession Code (Species) | Matched Distinct Peptide Sequences | Coverage (%) |
|---|---|---|---|---|---|
| NK-CTX | 201.05 | Cytotoxin 3 | P01446 (N. kaouthia) | LIPLAYK LIPLAYKTCPAGK LVPLFYK MFMVAAPK MFMVATPK MFMVSNK MFMVSNKTVPVK NSLLVKYVCCNTDR SSLLVK TCPAGKNLCYK YVCCNTDR GCIDACPK GCIDACPKNSLLVK | 92.7 |
| NS-CTX | 252.30 | Cytotoxin 1 | A0A7T7DMY7 (N. sumatrana) | LKCNKLVPLFYK CNKLVPLFYK DLCYK LIPLAYK LVPLFYK LVPLFYKTCPAGK MYMVATPK MYMVATPKVPVK NSLLVK RGCIDVCPK SSLLVK SSLLVKYVCCNTDR YVCCNTDR GCIDVCPK GCIDVCPKSSLLVK | 93.3 |
| NA-CTX | 208.83 | Cytotoxin 3 | P60301 (N. atra) | CNKLVPLFYK LVPLFYK LVPLFYKTCPAGK MFMVAAPK MFMVATPK MFMVSNK MYMVATPK NSLLVK RGCIDVCPK SSLLVK SSLLVKYVCCNTDR YVCCNTDR GCIDVCPK | 85.0 |
| Venom/Toxin | IC50 (Untreated) a (μg/mL) | NkMAV | SAV | NBAV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (Treated) b (μg/mL) | Antivenom Concentration (μg/mL) | Pc c (mg/g) | IC50 (Treated) b (μg/mL) | Antivenom Concentration (μg/mL) | Pc c (mg/g) | IC50 (Treated) b (μg/mL) | Antivenom Concentration (μg/mL) | Pc c (mg/g) | ||
| N. kaouthia (Thailand) | 47.40 ± 6.50 | 69.17 ± 8.39 | 125 | 174.16 | 82.17 ± 0.85 | 125 | 278.16 | 64.20 ± 7.00 | 125 | 134.40 |
| N. sumatrana (Malaysia) | 28.83 ± 2.86 | 37.47 ± 2.77 | 500 | 17.28 | 41.50 ± 1.25 | 500 | 25.34 | 36.50 ± 2.95 | 500 | 15.34 |
| N. atra (Taiwan) | 26.87 ± 8.86 | 49.60 ± 8.47 | 500 | 45.46 | 61.80 ± 6.73 | 500 | 69.86 | 43.53 ± 6.48 | 500 | 33.32 |
| NK-CTX | 32.90 ± 3.11 | 39.50 ± 2.40 | 500 | 13.20 | 38.23 ± 1.48 | 125 | 42.64 | 35.17 ± 7.10 | 500 | 4.54 |
| NS-CTX | 20.90 ± 3.73 | 39.33 ± 2.82 | 500 | 36.86 | 45.77 ± 9.40 | 500 | 49.74 | 36.10 ± 4.08 | 500 | 30.40 |
| NA-CTX | 13.17 ± 2.72 | 24.87 ± 1.36 | 500 | 23.40 | 31.50 ± 2.71 | 500 | 36.66 | 24.97 ± 1.95 | 500 | 23.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chong, H.P.; Tan, K.Y.; Liu, B.-S.; Sung, W.-C.; Tan, C.H. Cytotoxicity of Venoms and Cytotoxins from Asiatic Cobras (Naja kaouthia, Naja sumatrana, Naja atra) and Neutralization by Antivenoms from Thailand, Vietnam, and Taiwan. Toxins 2022, 14, 334. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins14050334
Chong HP, Tan KY, Liu B-S, Sung W-C, Tan CH. Cytotoxicity of Venoms and Cytotoxins from Asiatic Cobras (Naja kaouthia, Naja sumatrana, Naja atra) and Neutralization by Antivenoms from Thailand, Vietnam, and Taiwan. Toxins. 2022; 14(5):334. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins14050334
Chicago/Turabian StyleChong, Ho Phin, Kae Yi Tan, Bing-Sin Liu, Wang-Chou Sung, and Choo Hock Tan. 2022. "Cytotoxicity of Venoms and Cytotoxins from Asiatic Cobras (Naja kaouthia, Naja sumatrana, Naja atra) and Neutralization by Antivenoms from Thailand, Vietnam, and Taiwan" Toxins 14, no. 5: 334. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins14050334