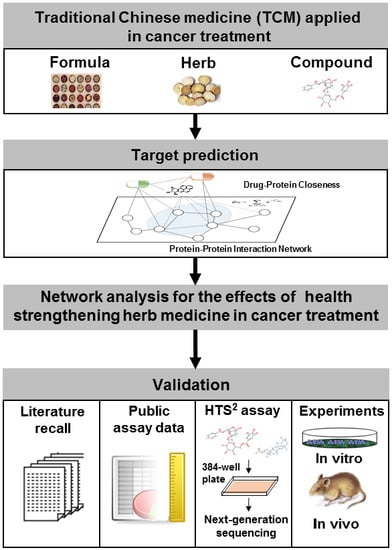
Network Pharmacology to Unveil the Biological Basis of Health-Strengthening Herbal Medicine in Cancer Treatment
Abstract
:1. Introduction
2. Results
2.1. The Prediction and Examination of Potential Targets of by Literature Mining and the HTS2 Assay
2.2. Target Prediction and Assay Results Indicate that the Two Types of TCM Herbs May Regulate Several Key Biological Processes in Cancer Treatment, Including Antitumor and Immune Modulation
2.3. HTS2 Assay Results Show that Health-Strengthening Medicine May Regulate Tumor Immunity Via Promoting NK Cell Activity and Tumor Cell Antigen Presentation
2.4. Target Prediction and HTS2 Assay Results Show that Compounds in the Same Herb May Exhibit Different Patterns in Modulating Antitumor or Immune Processes
2.5. Prediction and Assay Results Imply that Compounds in One Health-Strengthening Herb or a Single Compound May Exert Antitumor and Immune-Related Functions Simultaneously for Cancer Therapy
3. Discussion
4. Materials and Methods
4.1. TCM Compounds Data Preparation
4.2. Analysis Workflow Based on Network Pharmacology
4.3. Literature Mining
4.4. Target Prediction for the TCM Compounds Applied in Cancer Treatment
4.5. KEGG Pathway Enrichment Analysis
4.6. Chemical Space Analysis
4.7. Network Visualization
4.8. Cell Culture
4.9. The HTS2 Assay
4.9.1. Selection and Preparation of the TCM Compounds for HTS2
4.9.2. The Gene Selection and Probe Design of the HTS2 Assay
4.9.3. HTS2 Data Processing
4.10. Cell Viability Assay
4.11. Animal Studies
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Wang, S.; Zhang, Y.; Fan, H.T.; Lin, H.S. Traditional Chinese medicine and cancer: History, present situation, and development. Thorac. Cancer 2015, 6, 561–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, W.; Peng, L.; Shi, L.; Li, S. Traditional Chinese patent medicines for cancer treatment in China: A nationwide medical insurance data analysis. Oncotarget 2015, 6, 38283–38295. [Google Scholar]
- Qi, F.; Zhao, L.; Zhou, A.; Zhang, B.; Li, A.; Wang, Z.; Han, J. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. Biosci. Trends 2015, 9, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Chien, T.J.; Liu, C.Y.; Lu, R.H.; Kuo, C.W.; Lin, Y.C.; Hsu, C.H. Therapeutic efficacy of Traditional Chinese medicine, “Kuan-Sin-Yin”, in patients undergoing chemotherapy for advanced colon cancer—A controlled trial. Complement. Ther. Med. 2016, 29, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Parekh, H.S.; Liu, G.; Wei, M.Q. A new dawn for the use of traditional Chinese medicine in cancer therapy. Mol. Cancer 2009, 8, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Y.; Bai, X.Y.; Wang, C.H. Traditional Chinese medicine: A treasured natural resource of anticancer drug research and development. Am. J. Chin. Med. 2014, 42, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Human Resources and Social Security of the People’s Republic of China. China’s National Basic Medical Insurance, Work Injury Insurance and Maternity Insurance Drugs Catalog; (2017 Edition); China Labour and Social Security Publishing House: Beijing, China, 2017. [Google Scholar]
- Xiao, H.; Yang, J. Immune Enhancing Effect of Modified Sijunzi Decoction on Patients with Colorectal Cancer Undergoing Chemotherapy. Chin. J. Integr. Tradit. West. Med. 2011, 31, 164–167. [Google Scholar]
- Zhang, Y.; Guo, L.L.; Zhao, S.P. Effect of Shenqi Fuzheng Injection combined with chemotherapy in treating colorectal cancer. Zhongguo Zhong XI Yi Jie He Za Zhi 2010, 30, 280–282. [Google Scholar] [PubMed]
- Chen, Y.; Gan, L.; Zheng, W.; Wang, C.H.; Cheng, D.H. Effect of Shenqi Fuzheng Injection Combined with Chemotherapy in Treating Cancer Breast Cancer. Clin. J. Tradit. Chin. Med. 2016, 08, 1120–1122. [Google Scholar]
- Zhang, H.; Zhi-Xin, S.U. Curative effect of Shenling Baizhu powder on chemotherapy-induced toxicity in advanced gastric cancer. J. Tradit. Chin. Med. Univ. Hunan 2008. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, W.J. Clinical Observation the Recurrence of Colon Cancer after Treating with BuZhong YiQiTang Combined with Chemotherapy. West. J. Trad. Chin. Med. 2011, 24, 73–74. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, H. Buzhong Yiqi Decoction Relieve Toxic and Adverse Reactions of Advanced Lung Cancer Taking Chemotherapy. J. Zhejiang Univ. Tradit. Chin. Med. 2008, 32, 220–221. [Google Scholar]
- Zhu, X.; Chen, Y.; Zhong, X.; Zhang, X.Z.; Liu, J.H.; He, M. The protective effect of Shenqi Fuzheng Injection on immune function of BALB/c mice after chemo—therapy. Chin. J. Immunol. 2006, 22, 925–928. [Google Scholar]
- Fan, Y.; Dechuan, L.I.; Xinya, X.U. Study on Effect of Danggui Buxue Decoction Combined with Chemotherapy for Advanced Colorectal Cancer Patients with Immune Function. Chin. Arch. Tradit. Chin. Med. 2013. [Google Scholar] [CrossRef]
- Bao, S.Z.; Zhang, A.Q.; Sun, Z.D. Effect of Huangqijianzhong Decoction on Immune Function and Cyclin D1 Gene Expression in Mice with Lung Cancer of Spleen Qi Deficiency Syndrome. J. Emerg. Tradit. Chin. Med. 2012, 7, 032. [Google Scholar]
- Liang, X.; Li, H.; Li, S. A novel network pharmacology approach to analyse traditional herbal formulae: The Liu-Wei-Di-Huang pill as a case study. Mol. BioSyst. 2014, 10, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, S.Y.; Fang, Z.Q.; Guan, D.Y.; Wu, Z.H.; Gao, B.F. Study on the Mechanism of JianpiYiqi Decoction Regulating DEN- induced Hepatocarcinogenesis in Rats by Bioinformatic Analysis. Lishizhen Med. Mater. Med. Res. 2014, 25, 1765–1768. [Google Scholar]
- Yin, Y.; Feng, L.; Zhou, L.; Li, J.; Gao, Y.; Wang, N.J.; Yu, J.H.; Jiang, Z.L.; He, S.Q.; Lu, D.R.; et al. Effects of Yishengukang decoction on expression of bone-specific alkaline phosphatase, carboxyterminal propeptide of type I. procollagen, and carboxyterminal cross-linked telepeptide of type collagen in malignant tumor patients with bone metastasis. West. J. Trad. Chin. Med. 2017, 37, 30–34. [Google Scholar]
- Hsiao, W.L.; Liu, L. The role of traditional Chinese herbal medicines in cancer therapy--from TCM theory to mechanistic insights. Plant. Med. 2010, 76, 1118. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.; Wang, X.; Xu, N.; Zhang, H.M.; Xu, H.X. Application of proteomics to determine the mechanism of action of traditional Chinese medicine remedies. J. Ethnopharmacol. 2014, 155, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.C.; Xiao, P.G. Network pharmacology: A Rosetta Stone for traditional Chinese medicine. Drug Dev. Res. 2014, 75, 299. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhou, Q.; Ma, W.; Wei, D.Q. Exploring the Ligand-Protein Networks in Traditional Chinese Medicine: Current Databases, Methods, and Applications. Evid. Based Complement. Altern. Med. 2013, 2013, 227–257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, S. Network-Based Relating Pharmacological and Genomic Spaces for Drug Target Identification. PLoS ONE 2010, 5, e11764. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Nie, Q.; MacLean, A.; Li, Y.; Lei, J.; Li, S. Multiscale modeling of inflammation-induced tumorigenesis reveals competing oncogenic and onco-protective roles for inflammation. Cancer Research 2017, 77, 6429–6441. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Li, H.; Tian, G.; Li, S. Dynamic microbe and molecule networks in a mouse model of colitis-associated colorectal cancer. Sci. Rep. 2014, 4, 4985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Ma, T.; Gu, J.; Liang, X.; Li, S. Imbalanced network biomarkers for traditional Chinese medicine Syndrome in gastritis patients. Sci. Rep. 2013, 3, 1543. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Mapping ancient remedies: Applying a network approach to traditional Chinese medicine. Science 2015, 350, S72–S74. [Google Scholar]
- Hopkins, A.L. Network pharmacology:the next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, X.; Li, Y.; Wu, M.; Wang, S.; Li, S. Matrine is identified as a novel macropinocytosis inducer by a network target approach. Front. Pharmacol. 2018, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Macarron, R.; Banks, M.N.; Bojanic, D.; Burns, D.J.; Cirovic, D.A.; Garyantes, T.; Green, D.V.S.; Hertzberg, R.P.; Janzen, W.P.; Paslay, J.W.; et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, H.; Wang, D.; Qiu, J.S.; Zhou, Y.; Li, X.Q.; Rosenfeld, M.G.; Ding, S.; Fu, S.D. Versatile pathway-centric approach based on high-throughput sequencing to anticancer drug discovery. Proc. Nat. Acad. Sci. USA 2012, 109, 4609–4614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes pathways diseases drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009, 37, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Betel, D.; Hogue, C.W.V. BIND: The biomolecular interaction network database. Nucleic Acids Res. 2003, 31, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2011, 40, D841–D846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2011, 40, D857–D861. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.R.; Jurisica, I. Online predicted human interaction database. Bioinformatics 2005, 21, 2076–2082. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Coordinators, N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2013, 41, D8. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; Overington, J.P. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Team, R.S. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015; Volume 42, Available online: http://www.rstudio.com (accessed on April 2018).
- Martin, E.J.; Blaney, J.M.; Siani, M.A.; Spellmeyer, D.C.; Wong, A.K.; Moos, W.H. Measuring diversity: Experimental design of combinatorial libraries for drug discovery. J. Med. Chem. 1995, 38, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albertsson, P.A.; Basse, P.H.; Hokland, M.; Goldfarb, R.H.; Nagelkerke, J.F.; Nannmark, U.; Kuppen, P.J.K. NK cells and the tumour microenvironment: Implications for NK-cell function and anti-tumour activity. Trends Immunol. 2003, 24, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.; Cantoni, C.; Pietra, G.; Mingari, M.C.; Moretta, L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur. J. Immunol. 2014, 44, 1582–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, E.; James, E. Antigen processing and immune regulation in the response to tumours. Immunology 2016, 150, 16–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerezo-Wallis, D.; Soengas, M. Understanding Tumor-Antigen Presentation in the New Era of Cancer Immunotherapy. Curr. Pharm. Des. 2016, 22, 6234–6250. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Song, Z.; Guo, Q.; Li, J. Synergistic Effect and Molecular Mechanisms of Traditional Chinese Medicine on Regulating Tumor Microenvironment and Cancer Cells. BioMed Res. Int. 2016, 2016, 1490738. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.A.; Wheat, J.M.; Currie, G.M. Cancer stem cells and the impact of Chinese herbs, isolates and other complementary medical botanicals: A review. J. Chin. Integr. Med. 2012, 10, 493–503. [Google Scholar] [CrossRef]
- Fan, H.; Lin, H. Research Status of Fuzheng Herbs for Tumor Treating. World Chin. Med. 2014, 9, 825–832. [Google Scholar]
- Xiong, L.; Tian, S.X. A concept of regulating tumor microenvironment immune and normalizing angiogenesis by Chinese medicine drug therapy for supporting zheng-qi to prop up root. Chin. J. Integr. Tradit. West. Med. 2010, 30, 201–204. [Google Scholar]
- Qi, Q.; Li, R.; Li, H.; Cao, Y.; Bai, M.; Fan, X.; Wang, S.; Zhang, B.; Li, S. Identification of the anti-tumor activity and mechanisms of nuciferine through a network pharmacology approach. Acta Pharmacol. Sin. 2016, 37, 963–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Yu, G.; Li, J.; Xiong, F. Short term therapeutic effect on treatment of postoperational large intestine carcinoma by Fupiyiwei decoction combined with chemotherapy and it’s effect on immune function. China J. Chin. Mater. Med. 2010, 35, 782–785. [Google Scholar]
- Huang, Y.S.; Shi, Z.M. Intervention effect of Feiji Recipe on immune escape of lung cancer. Chin. J. Integr. Tradit. West. Med. 2007, 27, 501–504. [Google Scholar]
- Madlener, S.; Illmer, C.; Horvath, Z.; Saiko, P.; Losert, A.; Herbacek, I.; Grusch, M.; Elford, H.L.; Krupitza, G.; Bernhaus, A.; et al. Gallic acid inhibits ribonucleotide reductase and cyclooxygenases in human HL-60 promyelocytic leukemia cells. Cancer Lett. 2007, 245, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Chen, Z.W.; Wu, Y.M. Antitumor activity of total paeony glycoside against human chronic myelocytic leukemia K562 cell lines in vitro and in vivo. Med. Oncol. 2012, 29, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237. [Google Scholar] [CrossRef] [PubMed]
- Gotwals, P.; Cameron, S.; Cipolletta, D.; Cremasco, V.; Crystal, A.; Hewes, B.; Mueller, B.; Quaratino, S.; Sabatos-Peyton, C.; Petruzzelli, L.; et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 2017, 17, 286. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Willett, P.; Winterman, V.; Bawden, D. Implementation of nearest-neighbor searching in an online chemical structure search system. J. Chem. Inf. Comput. Sci. 1986, 26, 36–41. [Google Scholar] [CrossRef]
- Su, S.B.; Jia, W.; Lu, A.; Li, S. Evidence-based ZHENG: A traditional Chinese medicine syndrome. Evid.-Based Complement. Altern. Med. 2012, 2012, 246538. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, L.Y.; Wang, P.; Dai, H.; Gao, S.; Wang, K. Tumor microenvironment varies under different TCMZHENG models correlates with treatment response to herbal medicine. Evid.-Based Complement. Altern. Med. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Li, S.; Li, L.; Lin, K.; Liu, X.; Wang, H.; Wang, H.; Wang, D. Chemical genomics reveals inhibition of breast cancer lung metastasis by Ponatinib via c-Jun. Protein Cell. 2018, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The MathWorks. MATLAB (2016a); The MathWorks Inc.: Natick, MA, USA, 2016. [Google Scholar]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 40: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.; Ideker, T. Cytoscape 28: New features for data integration network visualization. Bioinformatics 2010, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man, (OMIM), a knowledgebase of human genes genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Jiang, R.; Zhang, M.Q.; Li, S. Network-based global inference of human disease genes. Mol. Syst. Biol. 2008, 4, 189. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Hao, H.; Li, Y.; Li, S. Modularity-based credible prediction of disease genes and detection of disease subtypes on the phenotype-gene heterogeneous network. BMC Syst. Biol. 2011, 5, 79. [Google Scholar] [CrossRef] [PubMed]






| KEGG Pathway | Herb Type | Herb | Compounds | Enrichment p-value | DEG Number |
|---|---|---|---|---|---|
| Apoptosis | Fu-Zheng | Radix Angelicae sinensis (Shan Yao) | Batatasin IV Dioscin | 2.1 × 10−4 1.1 × 10−2 | 13 23 |
| Qu-Xie | Rhizoma Curcumae (E Zhu) | Curcumin Isocurcumenol | 1.7 × 10−3 1.7 × 10−3 | 11 31 | |
| vascular endothelial growth factor (VEGF) signaling pathway | Fu-Zheng | Fructus Schisandrae (Wu Wei Zi) | Schisanhenol Gomisin J | 3.2 × 10−3 3.2 × 10−3 | 14 7 |
| Qu-Xie | Radix et Rhizoma Rhei (Da Huang) | Chrysaron Rhein | 2.3 × 10−6 3.2 × 10−3 | 9 5 | |
| Cell cycle | Fu-Zheng | Fructus Ligustri lucidi (Nv Zhen Zi) | Ligustroflavone Specnuezhenide | 2.4 × 10−3 1.5 × 10−3 | 4 10 |
| Qu-Xie | Cortex Moutan (Gan Chan Pi) | Cinobufagin Telocinobufagin | 1.5 × 10−3 2.3 × 10−4 | 20 26 | |
| T cell receptor signaling pathway | Fu-Zheng | Poria (Fu Ling) | Pachymic Acid Poricoic Acid B | 8.7 × 10−7 7.8 × 10−5 | 5 8 |
| Qu-Xie | Cortex Magnoliae officinali (Huang Qin) | Baicalein Wogonin | 7.8 × 10−5 3.8 × 10−3 | 18 15 | |
| Toll-like receptor signaling pathway | Fu-Zheng | Radix Astragali (Huang Qi) | Astragaloside A Formononetin | 4.1 × 10−3 4.1 × 10−3 | 5 3 |
| Qu Xie | Venenum Bufonis (Chan Su) | Bufarenogin Cinobufagin | 4.1 × 10−3 4.1 × 10−3 | 15 20 | |
| Nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway | Fu-Zheng | Radix Ginseng (Ren Shen) | Ginsenoside Rh3 Protopanaxadiol | 4.9 × 10−5 4.9 × 10−5 | 23 15 |
| Qu-Xie | Fructus Bruceae (Ya Dan Zi) | Bruceantin Bruceine D | 1.6 × 10−3 3.0 × 10−4 | 7 14 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Wu, M.; Wang, H.; Li, S.; Wang, X.; Li, Y.; Wang, D.; Li, S. Network Pharmacology to Unveil the Biological Basis of Health-Strengthening Herbal Medicine in Cancer Treatment. Cancers 2018, 10, 461. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers10110461
Zheng J, Wu M, Wang H, Li S, Wang X, Li Y, Wang D, Li S. Network Pharmacology to Unveil the Biological Basis of Health-Strengthening Herbal Medicine in Cancer Treatment. Cancers. 2018; 10(11):461. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers10110461
Chicago/Turabian StyleZheng, Jiahui, Min Wu, Haiyan Wang, Shasha Li, Xin Wang, Yan Li, Dong Wang, and Shao Li. 2018. "Network Pharmacology to Unveil the Biological Basis of Health-Strengthening Herbal Medicine in Cancer Treatment" Cancers 10, no. 11: 461. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers10110461