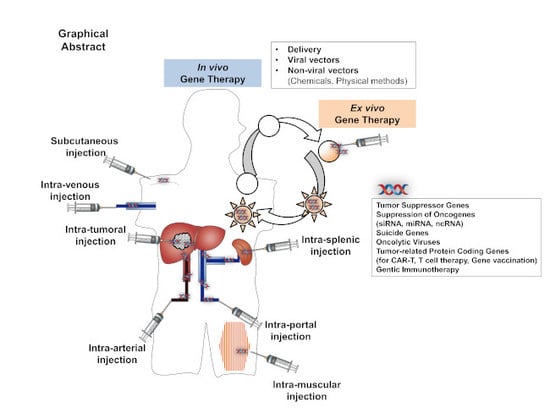
Gene Therapy for Liver Cancers: Current Status from Basic to Clinics
Abstract
:1. Introduction
1.1. Liver Cancers
1.1.1. Hepatocellular Carcinoma
1.1.2. Metastatic Liver Cancer
1.2. Gene Therapy
2. Gene Therapy for the Liver Cancers
2.1. Liver-Directed Gene Therapy
2.2. Target Genes for HCC Gene Therapy
2.2.1. Tumor Suppressor Genes
2.2.2. Oncogenes
2.2.3. Suicide Gene Therapy
2.2.4. Tumor Proteins
2.2.5. Genetic Immunotherapy
2.3. Gene Delivery Procedures
2.3.1. Viral Gene Delivery
2.3.2. Non-Viral Gene Delivery Using Chemicals
2.3.3. Non-Viral Gene Delivery Using Physical Methods
2.4. Clinical Trials
2.4.1. Ongoing Clinical Trials for Gene Therapy of Liver Cancers
2.4.2. Ongoing Clinical Trials for Gene-Based Diagnosis
3. Recent Progress
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dooley, J.S.; Lok, A.S.; Garcia-Tsao, G.; Pinzani, M. Sherlock’s Diseases of the Liver and Biliary System, 13th ed.; Wiley-Blackwell Science: Hoboken, NJ, USA, 2018. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Ogunwobi, O.O.; Harricharran, T.; Huaman, J.; Galuza, A.; Odumuwagun, O.; Tan, Y.; Ma, G.X.; Nguyen, M.T. Mechanisms of hepatocellular carcinoma progression. World J. Gastroenterol. 2019, 25, 2279–2293. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Goto, T.; Hirotsu, Y.; Moriyama, M.; Omata, M. Molecular Mechanisms Driving Progression of Liver Cirrhosis towards Hepatocellular Carcinoma in Chronic Hepatitis B and C Infections: A Review. Int. J. Mol. Sci. 2019, 20, 1358. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.; Gogineni, V.; Saeian, K. Epidemiology of primary and secondary liver cancers. Semin. Intervent. Radiol. 2006, 23, 47–63. [Google Scholar] [CrossRef]
- Bruix, J.; Reig, M.; Sherman, M. Evidence-Based Diagnosis, Staging, and Treatment of Patients with Hepatocellular Carcinoma. Gastroenterology 2016, 150, 835–853. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [CrossRef]
- Calderaro, J.; Ziol, M.; Paradis, V.; Zucman-Rossi, J. Molecular and histological correlations in liver cancer. J. Hepatol. 2019, 71, 616–630. [Google Scholar] [CrossRef]
- Elnaggar, M.H.; Abushouk, A.I.; Lamloum, H.M.; Benmelouka, A.; Moatamed, S.A.; Abd-Elmegeed, H.; Attia, S.; Samir, A.; Amr, N.; Johar, D.; et al. Nanomedicine As A Putative Approach For Active Targeting of Hepatocellular Carcinoma. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Llovet, J.M.; Montal, R.; Villanueva, A. Randomized trials and endpoints in advanced HCC: Role of PFS as a surrogate of survival. J. Hepatol. 2019, 70, 1262–1277. [Google Scholar] [CrossRef]
- Raja, A.; Park, I.; Haq, F.; Ahn, S.M. FGF19-FGFR4 Signaling in Hepatocellular Carcinoma. Cells 2019, 8, 536. [Google Scholar] [CrossRef] [PubMed]
- El Dika, I.; Khalil, D.N.; Abou-Alfa, G.K. Immune checkpoint inhibitors for hepatocellular carcinoma. Cancer 2019, 125, 3312–3319. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, S.; Zeng, S.; Shen, H. From bench to bed: The tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 396. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Rong, D.; Zhang, B.; Zheng, W.; Wang, X.; Chen, Z.; Tang, W. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: Challenges and opportunities. Mol. Cancer 2019, 18, 130. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, H.; Sun, B.; Karin, M. The Immunobiology of Hepatocellular Carcinoma in Humans and Mice: Basic Concepts and Therapeutic Implications. J. Hepatol. 2019. [Google Scholar] [CrossRef]
- Alaei-Mahabadi, B.; Bhadury, J.; Karlsson, J.W.; Nilsson, J.A.; Larsson, E. Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc. Natl. Acad. Sci. USA 2016, 113, 13768–13773. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef]
- Rao, C.V.; Asch, A.S.; Yamada, H.Y. Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. Carcinogenesis 2017, 38, 2–11. [Google Scholar] [CrossRef]
- Gillet, J.P.; Andersen, J.B.; Madigan, J.P.; Varma, S.; Bagni, R.K.; Powell, K.; Burgan, W.E.; Wu, C.P.; Calcagno, A.M.; Ambudkar, S.V.; et al. A Gene Expression Signature Associated with Overall Survival in Patients with Hepatocellular Carcinoma Suggests a New Treatment Strategy. Mol. Pharmacol. 2016, 89, 263–272. [Google Scholar] [CrossRef]
- Sun, W.; Shi, Q.; Zhang, H.; Yang, K.; Ke, Y.; Wang, Y.; Qiao, L. Advances in the techniques and methodologies of cancer gene therapy. Discov. Med. 2019, 27, 45–55. [Google Scholar]
- Wang, X.; Tai, Z.; Zhang, W.; Gao, S. Current status of gene therapy for hepatocellular carcinoma, with a focus on gene delivery approaches. Curr. Gene Ther. 2015, 15, 120–141. [Google Scholar] [CrossRef] [PubMed]
- Reghupaty, S.C.; Sarkar, D. Current Status of Gene Therapy in Hepatocellular Carcinoma. Cancers (Basel) 2019, 11, 1265. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef]
- High, K.A.; Roncarolo, M.G. Gene Therapy. N. Engl. J. Med. 2019, 381, 455–464. [Google Scholar] [CrossRef]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef]
- Karimian, A.; Azizian, K.; Parsian, H.; Rafieian, S.; Shafiei-Irannejad, V.; Kheyrollah, M.; Yousefi, M.; Majidinia, M.; Yousefi, B. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J. Cell. Physiol. 2019, 234, 12267–12277. [Google Scholar] [CrossRef]
- Chew, W.L.; Tabebordbar, M.; Cheng, J.K.; Mali, P.; Wu, E.Y.; Ng, A.H.; Zhu, K.; Wagers, A.J.; Church, G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 2016, 13, 868–874. [Google Scholar] [CrossRef]
- Alsaggara, M.; Liu, D. Liver-Targeted Gene and Cell Therapies: An Overview. In Gene Therapy and Cell Therapy through the Liver; Terai, S., Suda, T., Eds.; Springer: Tokyo, Japan, 2016. [Google Scholar]
- Yin, H.; Song, C.Q.; Dorkin, J.R.; Zhu, L.J.; Li, Y.; Wu, Q.; Park, A.; Yang, J.; Suresh, S.; Bizhanova, A.; et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016, 34, 328–333. [Google Scholar] [CrossRef]
- Meng, X.; Franklin, D.A.; Dong, J.; Zhang, Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014, 74, 7161–7167. [Google Scholar] [CrossRef]
- Erstad, D.J.; Fuchs, B.C.; Tanabe, K.K. Molecular signatures in hepatocellular carcinoma: A step toward rationally designed cancer therapy. Cancer 2018, 124, 3084–3104. [Google Scholar] [CrossRef] [PubMed]
- Anestopoulos, I.; Voulgaridou, G.P.; Georgakilas, A.G.; Franco, R.; Pappa, A.; Panayiotidis, M.I. Epigenetic therapy as a novel approach in hepatocellular carcinoma. Pharmacol. Ther. 2015, 145, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Quetglas, I.M.; Moeini, A.; Pinyol, R.; Llovet, J.M. Integration of genomic information in the clinical management of HCC. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Scaggiante, B.; Kazemi, M.; Pozzato, G.; Dapas, B.; Farra, R.; Grassi, M.; Zanconati, F.; Grassi, G. Novel hepatocellular carcinoma molecules with prognostic and therapeutic potentials. World J. Gastroenterol. 2014, 20, 1268–1288. [Google Scholar] [CrossRef]
- Berasain, C.; Avila, M.A. The EGFR signalling system in the liver: From hepatoprotection to hepatocarcinogenesis. J. Gastroenterol. 2014, 49, 9–23. [Google Scholar] [CrossRef]
- Xiao, H.; Tong, R.; Ding, C.; Lv, Z.; Du, C.; Peng, C.; Cheng, S.; Xie, H.; Zhou, L.; Wu, J.; et al. Gamma-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1alpha/VEGF pathways under hypoxic condition. Oncotarget 2015, 6, 2180–2192. [Google Scholar] [CrossRef]
- Wen, Y.; Zhou, X.; Lu, M.; He, M.; Tian, Y.; Liu, L.; Wang, M.; Tan, W.; Deng, Y.; Yang, X.; et al. Bclaf1 promotes angiogenesis by regulating HIF-1alpha transcription in hepatocellular carcinoma. Oncogene 2019, 38, 1845–1859. [Google Scholar] [CrossRef]
- Ni, J.Y.; Xu, L.F.; Wang, W.D.; Huang, Q.S.; Sun, H.L.; Chen, Y.T. Transarterial embolization combined with RNA interference targeting hypoxia-inducible factor-1alpha for hepatocellular carcinoma: A preliminary study of rat model. J. Cancer Res. Clin. Oncol. 2017, 143, 199–207. [Google Scholar] [CrossRef]
- Gu, D.L.; Chen, Y.H.; Shih, J.H.; Lin, C.H.; Jou, Y.S.; Chen, C.F. Target genes discovery through copy number alteration analysis in human hepatocellular carcinoma. World J. Gastroenterol. 2013, 19, 8873–8879. [Google Scholar] [CrossRef]
- Cornella, H.; Alsinet, C.; Sayols, S.; Zhang, Z.; Hao, K.; Cabellos, L.; Hoshida, Y.; Villanueva, A.; Thung, S.; Ward, S.C.; et al. Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology 2015, 148, 806–818. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Garrido, P.; Andersen, J.B. Genetic Optimization of Liver Cancer Therapy: A Patient-Derived Primary Cancer Cell-Based Model. Gastroenterology 2017, 152, 19–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.; Wang, Z.C.; Duan, M.; Lin, Y.H.; Zhou, X.Y.; Worthley, D.L.; Wang, X.Y.; Niu, G.; Xia, Y.; Deng, M.; et al. Cell Culture System for Analysis of Genetic Heterogeneity Within Hepatocellular Carcinomas and Response to Pharmacologic Agents. Gastroenterology 2017, 152, 232–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Electronic address: [email protected]; Cancer Genome Atlas Research Network: Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Shen, A.; Liu, S.; Yu, W.; Deng, H.; Li, Q. p53 gene therapy-based transarterial chemoembolization for unresectable hepatocellular carcinoma: A prospective cohort study. J. Gastroenterol. Hepatol. 2015, 30, 1651–1656. [Google Scholar] [CrossRef]
- Chawla, S.P.; Bruckner, H.; Morse, M.A.; Assudani, N.; Hall, F.L.; Gordon, E.M. A Phase I-II Study Using Rexin-G Tumor-Targeted Retrovector Encoding a Dominant-Negative Cyclin G1 Inhibitor for Advanced Pancreatic Cancer. Mol. Ther. Oncolytics 2019, 12, 56–67. [Google Scholar] [CrossRef] [Green Version]
- Santiago-Ortiz, J.L.; Schaffer, D.V. Adeno-associated virus (AAV) vectors in cancer gene therapy. J. Control. Release 2016, 240, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Huang, F.; Cai, H.; Zhong, S.; Liu, X.; Tan, W.S. Potent antitumor effect of TRAIL mediated by a novel adeno-associated viral vector targeting to telomerase activity for human hepatocellular carcinoma. J. Gene Med. 2008, 10, 518–526. [Google Scholar] [CrossRef]
- Ran, Z.; Chen, W.; Shang, J.; Li, X.; Nie, Z.; Yang, J.; Li, N. Clinicopathological and prognostic implications of polo-like kinase 1 expression in colorectal cancer: A systematic review and meta-analysis. Gene 2019, 721, 144097. [Google Scholar] [CrossRef]
- Fu, Z.; Wen, D. The Emerging Role of Polo-Like Kinase 1 in Epithelial-Mesenchymal Transition and Tumor Metastasis. Cancers (Basel) 2017, 9, 131. [Google Scholar] [CrossRef] [Green Version]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 2006, 6, 321–330. [Google Scholar] [CrossRef]
- Van Haele, M.; Moya, I.M.; Karaman, R.; Rens, G.; Snoeck, J.; Govaere, O.; Nevens, F.; Verslype, C.; Topal, B.; Monbaliu, D.; et al. YAP and TAZ Heterogeneity in Primary Liver Cancer: An Analysis of Its Prognostic and Diagnostic Role. Int. J. Mol. Sci. 2019, 20, 638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Lu, T.; Li, J.; Yang, R.; Hu, L.; Ye, Y.; Mao, F.; He, L.; Xu, J.; Wang, Z.; et al. The novel tumor suppressor IRF2BP2 regulates Hippo pathway in liver cancer via a feedback loop. Hepatology 2019. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhou, J.K.; Wang, K.; Chen, H.; Qin, S.; Liu, J.; Luo, M.; Chen, Y.; Jiang, J.; Zhou, L.; et al. PDLIM1 inhibits tumor metastasis through activating Hippo signaling in hepatocellular carcinoma. Hepatology 2019. [Google Scholar] [CrossRef] [PubMed]
- Rafatmanesh, A.; Behjati, M.; Mobasseri, N.; Sarvizadeh, M.; Mazoochi, T.; Karimian, M. The survivin molecule as a double-edged sword in cellular physiologic and pathologic conditions and its role as a potential biomarker and therapeutic target in cancer. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Namgung, Y.; Kim, S.Y.; Kim, I. Down-regulation of Survivin by BIX-01294 Pretreatment Overcomes Resistance of Hepatocellular Carcinoma Cells to TRAIL. Anticancer Res. 2019, 39, 3571–3578. [Google Scholar] [CrossRef]
- Sehgal, A.; Vaishnaw, A.; Fitzgerald, K. Liver as a target for oligonucleotide therapeutics. J. Hepatol. 2013, 59, 1354–1359. [Google Scholar] [CrossRef] [Green Version]
- Berraondo, P.; Martini, P.G.V.; Avila, M.A.; Fontanellas, A. Messenger RNA therapy for rare genetic metabolic diseases. Gut 2019, 68, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Hajiasgharzadeh, K.; Somi, M.H.; Shanehbandi, D.; Mokhtarzadeh, A.; Baradaran, B. Small interfering RNA-mediated gene suppression as a therapeutic intervention in hepatocellular carcinoma. J. Cell. Physiol. 2019, 234, 3263–3276. [Google Scholar] [CrossRef]
- Scarabel, L.; Perrone, F.; Garziera, M.; Farra, R.; Grassi, M.; Musiani, F.; Russo Spena, C.; Salis, B.; De Stefano, L.; Toffoli, G.; et al. Strategies to optimize siRNA delivery to hepatocellular carcinoma cells. Expert Opin. Drug Deliv. 2017, 14, 797–810. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Nakao, K.; Miyaaki, H.; Ichikawa, T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J. Gastroenterol. 2014, 49, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Lanzafame, M.; Bianco, G.; Terracciano, L.M.; Ng, C.K.Y.; Piscuoglio, S. The Role of Long Non-Coding RNAs in Hepatocarcinogenesis. Int. J. Mol. Sci. 2018, 19, 682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, L.; Tang, Q.; Li, G.; Chen, K. Long non-coding RNAs as biomarkers and therapeutic targets: Recent insights into hepatocellular carcinoma. Life Sci. 2017, 191, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, S.; Liu, Y.; Meng, Z.; Chen, F. Low lncRNA ZNF385DAS2 expression and its prognostic significance in liver cancer. Oncol. Rep. 2019, 42, 1110–1124. [Google Scholar] [PubMed]
- Huang, J.L.; Zheng, L.; Hu, Y.W.; Wang, Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis 2014, 35, 507–514. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xu, X. Biological functions and clinical applications of exosomal non-coding RNAs in hepatocellular carcinoma. Cell. Mol. Life Sci. 2019, 76, 4203–4219. [Google Scholar] [CrossRef]
- Springer, A.D.; Dowdy, S.F. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther. 2018, 28, 109–118. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Duzgunes, N. Origins of Suicide Gene Therapy. Methods Mol. Biol. 2019, 1895, 1–9. [Google Scholar]
- Sangro, B.; Mazzolini, G.; Ruiz, M.; Ruiz, J.; Quiroga, J.; Herrero, I.; Qian, C.; Benito, A.; Larrache, J.; Olague, C.; et al. A phase I clinical trial of thymidine kinase-based gene therapy in advanced hepatocellular carcinoma. Cancer Gene Ther. 2010, 17, 837–843. [Google Scholar] [CrossRef]
- Kemeny, N.; Brown, K.; Covey, A.; Kim, T.; Bhargava, A.; Brody, L.; Guilfoyle, B.; Haag, N.P.; Karrasch, M.; Glasschroeder, B.; et al. Phase I, open-label, dose-escalating study of a genetically engineered herpes simplex virus, NV1020, in subjects with metastatic colorectal carcinoma to the liver. Hum. Gene Ther. 2006, 17, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Wills, K.N.; Huang, W.M.; Harris, M.P.; Machemer, T.; Maneval, D.C.; Gregory, R.J. Gene therapy for hepatocellular carcinoma: Chemosensitivity conferred by adenovirus-mediated transfer of the HSV-1 thymidine kinase gene. Cancer Gene Ther. 1995, 2, 191–197. [Google Scholar] [PubMed]
- Yamada, T.; Hamano, Y.; Hasegawa, N.; Seo, E.; Fukuda, K.; Yokoyama, K.K.; Hyodo, I.; Abei, M. Oncolytic Virotherapy and Gene Therapy Strategies for Hepatobiliary Cancers. Curr. Cancer Drug Targets 2018, 18, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Harrington, K.; Freeman, D.J.; Kelly, B.; Harper, J.; Soria, J.C. Optimizing oncolytic virotherapy in cancer treatment. Nat. Rev. Drug Discov. 2019, 18, 689–706. [Google Scholar] [CrossRef]
- Lai, Y.H.; Lin, C.C.; Chen, S.H.; Tai, C.K. Tumor-specific suicide gene therapy for hepatocellular carcinoma by transcriptionally targeted retroviral replicating vectors. Gene Ther. 2015, 22, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Yu, Y.P.; Zuo, Z.H.; Nelson, J.B.; Michalopoulos, G.K.; Monga, S.; Liu, S.; Tseng, G.; Luo, J.H. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nat. Biotechnol. 2017, 35, 543–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.; Gao, A.; Zhang, R.; Ma, H.; Shen, H.; Hu, Q.; Zhang, H.; Zhao, M.; Lan, X.; Liu, K. Use of a Novel Integrase-Deficient Lentivirus for Targeted Anti-Cancer Therapy with Survivin Promoter-Driven Diphtheria Toxin A. Medicine 2015, 94, e1301. [Google Scholar] [CrossRef]
- Hanna, N.; Ohana, P.; Konikoff, F.M.; Leichtmann, G.; Hubert, A.; Appelbaum, L.; Kopelman, Y.; Czerniak, A.; Hochberg, A. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. 2012, 19, 374–381. [Google Scholar] [CrossRef]
- Scaiewicz, V.; Sorin, V.; Fellig, Y.; Birman, T.; Mizrahi, A.; Galula, J.; Abu-Lail, R.; Shneider, T.; Ohana, P.; Buscail, L.; et al. Use of H19 Gene Regulatory Sequences in DNA-Based Therapy for Pancreatic Cancer. J. Oncol. 2010, 2010. [Google Scholar] [CrossRef] [Green Version]
- Mizrahi, A.; Czerniak, A.; Levy, T.; Amiur, S.; Gallula, J.; Matouk, I.; Abu-lail, R.; Sorin, V.; Birman, T.; de Groot, N.; et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 2009, 7, 69. [Google Scholar] [CrossRef] [Green Version]
- Amit, D.; Hochberg, A. Development of targeted therapy for a broad spectrum of cancers (pancreatic cancer, ovarian cancer, glioblastoma and HCC) mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Int. J. Clin. Exp. Med. 2012, 5, 296–305. [Google Scholar] [PubMed]
- Sidi, A.A.; Ohana, P.; Benjamin, S.; Shalev, M.; Ransom, J.H.; Lamm, D.; Hochberg, A.; Leibovitch, I. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J. Urol. 2008, 180, 2379–2383. [Google Scholar] [CrossRef] [PubMed]
- Montalbano, M.; Georgiadis, J.; Masterson, A.L.; McGuire, J.T.; Prajapati, J.; Shirafkan, A.; Rastellini, C.; Cicalese, L. Biology and function of glypican-3 as a candidate for early cancerous transformation of hepatocytes in hepatocellular carcinoma (Review). Oncol. Rep. 2017, 37, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Jiang, X.; Chen, S.; Lai, Y.; Wei, X.; Li, B.; Lin, S.; Wang, S.; Wu, Q.; Liang, Q.; et al. Anti-GPC3-CAR T Cells Suppress the Growth of Tumor Cells in Patient-Derived Xenografts of Hepatocellular Carcinoma. Front. Immunol. 2016, 7, 690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flecken, T.; Schmidt, N.; Hild, S.; Gostick, E.; Drognitz, O.; Zeiser, R.; Schemmer, P.; Bruns, H.; Eiermann, T.; Price, D.A.; et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology 2014, 59, 1415–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dauch, D.; Rudalska, R.; Cossa, G.; Nault, J.C.; Kang, T.W.; Wuestefeld, T.; Hohmeyer, A.; Imbeaud, S.; Yevsa, T.; Hoenicke, L.; et al. A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat. Med. 2016, 22, 744–753. [Google Scholar] [CrossRef]
- Kanwal, F.; Singal, A.G. Surveillance for Hepatocellular Carcinoma: Current Best Practice and Future Direction. Gastroenterology 2019, 157, 54–64. [Google Scholar] [CrossRef] [Green Version]
- Wong, R.J.; Ahmed, A.; Gish, R.G. Elevated alpha-fetoprotein: Differential diagnosis—Hepatocellular carcinoma and other disorders. Clin. Liver Dis. 2015, 19, 309–323. [Google Scholar] [CrossRef]
- Butterfield, L.H.; Economou, J.S.; Gamblin, T.C.; Geller, D.A. Alpha fetoprotein DNA prime and adenovirus boost immunization of two hepatocellular cancer patients. J. Transl. Med. 2014, 12, 86. [Google Scholar] [CrossRef] [Green Version]
- Bakalakos, E.A.; Burak, W.E., Jr.; Young, D.C.; Martin, E., Jr. Is carcino-embryonic antigen useful in the follow-up management of patients with colorectal liver metastases? Am. J. Surg. 1999, 177, 2–6. [Google Scholar] [CrossRef]
- Rizeq, B.; Zakaria, Z.; Ouhtit, A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci. 2018, 109, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Morse, M.A.; Niedzwiecki, D.; Marshall, J.L.; Garrett, C.; Chang, D.Z.; Aklilu, M.; Crocenzi, T.S.; Cole, D.J.; Dessureault, S.; Hobeika, A.C.; et al. A randomized phase II study of immunization with dendritic cells modified with poxvectors encoding CEA and MUC1 compared with the same poxvectors plus GM-CSF for resected metastatic colorectal cancer. Ann. Surg. 2013, 258, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Gordeeva, O. Cancer-testis antigens: Unique cancer stem cell biomarkers and targets for cancer therapy. Semin. Cancer Biol. 2018, 53, 75–89. [Google Scholar] [CrossRef]
- Patel, S.V.; Khan, D.A. Adverse Reactions to Biologic Therapy. Immunol. Allergy Clin. 2017, 37, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Lam, M.G. Delivery approaches of gene therapy in hepatocellular carcinoma. AntiCancer Res. 2013, 33, 4711–4718. [Google Scholar] [PubMed]
- Wang, Y.; Du, H.; Zhai, G. Recent advances in active hepatic targeting drug delivery system. Curr. Drug Targets 2014, 15, 573–599. [Google Scholar] [CrossRef]
- Kamimura, K.; Suda, T.; Zhang, G.; Liu, D. Advances in gene delivery systems. Pharm. Med. 2011, 25, 293–306. [Google Scholar] [CrossRef]
- Kamimura, K.; Liu, D. Physical approaches for nucleic acid delivery to liver. AAPS J. 2008, 10, 589–595. [Google Scholar] [CrossRef]
- Kamimura, K.; Suda, T.; Kanefuji, T.; Yokoo, T.; Abe, H.; Kobayashi, Y.; Aoyagi, Y.; Liu, D. Image-Guided Hydrodynamic Gene Delivery to the Liver: Toward Clinical Applications. In Gene Therapy and Cell Therapy through the Liver; Terai, S., Suda, T., Eds.; Springer: Tokyo, Japan, 2016; Chapter 8. [Google Scholar]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Rittie, L.; Athanasopoulos, T.; Calero-Garcia, M.; Davies, M.L.; Dow, D.J.; Howe, S.J.; Morrison, A.; Ricciardelli, I.; Saudemont, A.; Jespers, L.; et al. The Landscape of Early Clinical Gene Therapies outside of Oncology. Mol. Ther. 2019. [Google Scholar] [CrossRef] [PubMed]
- van der Laan, L.J.; Wang, Y.; Tilanus, H.W.; Janssen, H.L.; Pan, Q. AAV-mediated gene therapy for liver diseases: The prime candidate for clinical application? Expert Opin. Biol. Ther. 2011, 11, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Dismuke, D.; Samulski, R.J. Hepatic gene therapy using lentiviral vectors: Has safety been established? Hepatology 2013, 58, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Vetrini, F.; Ng, P. Liver-directed gene therapy with helper-dependent adenoviral vectors: Current state of the art and future challenges. Curr. Pharm. Des. 2011, 17, 2488–2499. [Google Scholar] [CrossRef]
- Sweeney, N.P.; Meng, J.; Patterson, H.; Morgan, J.E.; McClure, M. Delivery of large transgene cassettes by foamy virus vector. Sci. Rep. 2017, 7, 8085. [Google Scholar] [CrossRef] [Green Version]
- Uchida, H.; Hamada, H.; Nakano, K.; Kwon, H.; Tahara, H.; Cohen, J.B.; Glorioso, J.C. Oncolytic Herpes Simplex Virus Vectors Fully Retargeted to Tumor- Associated Antigens. Curr. Cancer Drug Targets 2018, 18, 162–170. [Google Scholar] [CrossRef]
- Liu, J.; Jaijyan, D.K.; Tang, Q.; Zhu, H. Promising Cytomegalovirus-Based Vaccine Vector Induces Robust CD8(+) T-Cell Response. Int. J. Mol. Sci. 2019, 20, 4457. [Google Scholar] [CrossRef] [Green Version]
- Atchison, R.W.; Casto, B.C.; Hammon, W.M. Adenovirus-associated defective virus particles. Science 1965, 149, 754–756. [Google Scholar] [CrossRef]
- Wang, Y.G.; Huang, P.P.; Zhang, R.; Ma, B.Y.; Zhou, X.M.; Sun, Y.F. Targeting adeno-associated virus and adenoviral gene therapy for hepatocellular carcinoma. World J. Gastroenterol. 2016, 22, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Chandler, R.J.; Tarasenko, T.N.; Cusmano-Ozog, K.; Sun, Q.; Sutton, V.R.; Venditti, C.P.; McGuire, P.J. Liver-directed adeno-associated virus serotype 8 gene transfer rescues a lethal murine model of citrullinemia type 1. Gene Ther. 2013, 20, 1188. [Google Scholar] [CrossRef] [Green Version]
- Nathwani, A.C.; Tuddenham, E.G.; Rangarajan, S.; Rosales, C.; McIntosh, J.; Linch, D.C.; Chowdary, P.; Riddell, A.; Pie, A.J.; Harrington, C.; et al. Adenovirus-Associated Virus Vector-Mediated Gene Transfer in Hemophilia, B. N. Engl. J. Med. 2011, 365, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Peyvandi, F.; Garagiola, I. Clinical advances in gene therapy updates on clinical trials of gene therapy in hemophilia. Haemophilia 2019, 25, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lu, Y.; Witek, R.P.; Chang, L.J.; Campbell-Thompson, M.; Jorgensen, M.; Petersen, B.; Song, S. Ex vivo transduction and transplantation of bone marrow cells for liver gene delivery of alpha1-antitrypsin. Mol. Ther. 2010, 18, 1553–1558. [Google Scholar] [CrossRef]
- Suhy, D.A.; Kao, S.C.; Mao, T.; Whiteley, L.; Denise, H.; Souberbielle, B.; Burdick, A.D.; Hayes, K.; Wright, J.F.; Lavender, H.; et al. Safe, long-term hepatic expression of anti-HCV shRNA in a nonhuman primate model. Mol. Ther. 2012, 20, 1737–1749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; Chang, J.C.; Xu, S.M.; Kan, Y.W. Selective killing of AFP-positive hepatocellular carcinoma cells by adeno-associated virus transfer of the herpes simplex virus thymidine kinase gene. Hum. Gene Ther. 1996, 7, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Bell, P.; Gao, G.; Haskins, M.E.; Wang, L.; Sleeper, M.; Wang, H.; Calcedo, R.; Vandenberghe, L.H.; Chen, S.J.; Weisse, C.; et al. Evaluation of adeno-associated viral vectors for liver-directed gene transfer in dogs. Hum. Gene Ther. 2011, 22, 985–997. [Google Scholar] [CrossRef] [Green Version]
- Brunetti-Pierri, N.; Liou, A.; Patel, P.; Palmer, D.; Grove, N.; Finegold, M.; Piccolo, P.; Donnachie, E.; Rice, K.; Beaudet, A.; et al. Balloon Catheter Delivery of Helper-dependent Adenoviral Vector Results in Sustained, Therapeutic hFIX Expression in Rhesus Macaques. Mol. Ther. 2012, 20, 1863–1870. [Google Scholar] [CrossRef] [Green Version]
- Sobrevals, L.; Enguita, M.; Rodriguez, C.; Gonzalez-Rojas, J.; Alzaguren, P.; Razquin, N.; Prieto, J.; Fortes, P. AAV vectors transduce hepatocytes in vivo as efficiently in cirrhotic as in healthy rat livers. Gene Ther. 2012, 19, 411–417. [Google Scholar] [CrossRef]
- Sabatino, D.E.; Lange, A.M.; Altynova, E.S.; Sarkar, R.; Zhou, S.; Merricks, E.P.; Franck, H.G.; Nichols, T.C.; Arruda, V.R.; Kazazian Jr, H. Efficacy and safety of long-term prophylaxis in severe hemophilia A dogs following liver gene therapy using AAV vectors. Mol. Ther. 2011, 19, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Penuelas, I.; Mazzolini, G.; Boan, J.F.; Sangro, B.; Marti-Climent, J.; Ruiz, M.; Ruiz, J.; Satyamurthy, N.; Qian, C.; Barrio, J.R.; et al. Positron emission tomography imaging of adenoviral-mediated transgene expression in liver cancer patients. Gastroenterology 2005, 128, 1787–1795. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Ma, N.; Steinhoff, G. Non-viral gene delivery methods. Curr. Pharm. Biotechnol. 2013, 14, 46–60. [Google Scholar] [PubMed]
- Pal Singh, P.; Vithalapuram, V.; Metre, S.; Kodipyaka, R. Lipoplex-based therapeutics for effective oligonucleotide delivery: A compendious review. J. Liposome Res. 2019, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, R.A.; Serra, A.; Coelho, J.F.J.; Faneca, H. Poly (beta-amino ester)-based gene delivery systems: From discovery to therapeutic applications. J. Control. Release 2019, 310, 155–187. [Google Scholar] [CrossRef] [PubMed]
- Bolhassani, A. Improvements in chemical carriers of proteins and peptides. Cell Biol. Int. 2019, 43, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic. Acid Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [Green Version]
- Varshosaz, J.; Farzan, M. Nanoparticles for targeted delivery of therapeutics and small interfering RNAs in hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 12022–12041. [Google Scholar] [CrossRef]
- Hashida, M.; Nishikawa, M.; Yamashita, F.; Takakura, Y. Cell-specific delivery of genes with glycosylated carriers. Adv. Drug Deliv. Rev. 2001, 52, 187–196. [Google Scholar] [CrossRef]
- Adrian, J.E.; Poelstra, K.; Scherphof, G.L.; Meijer, D.K.; van Loenen-Weemaes, A.M.; Reker-Smit, C.; Morselt, H.W.; Zwiers, P.; Kamps, J.A. Effects of a new bioactive lipid-based drug carrier on cultured hepatic stellate cells and liver fibrosis in bile duct-ligated rats. J. Pharmacol. Exp. Ther. 2007, 321, 536–543. [Google Scholar] [CrossRef] [Green Version]
- Puxbaum, V.; Nimmerfall, E.; Bauerl, C.; Taub, N.; Blaas, P.M.; Wieser, J.; Mikula, M.; Mikulits, W.; Ng, K.M.; Yeoh, G.C.; et al. M6P/IGF2R modulates the invasiveness of liver cells via its capacity to bind mannose 6-phosphate residues. J. Hepatol. 2012, 57, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Zakeri, A.; Kouhbanani, M.A.J.; Beheshtkhoo, N.; Beigi, V.; Mousavi, S.M.; Hashemi, S.A.R.; Karimi Zade, A.; Amani, A.M.; Savardashtaki, A.; Mirzaei, E.; et al. Polyethylenimine-based nanocarriers in co-delivery of drug and gene: A developing horizon. Nano Rev. Exp. 2018, 9, 1488497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamboni, C.G.; Kozielski, K.L.; Vaughan, H.J.; Nakata, M.M.; Kim, J.; Higgins, L.J.; Pomper, M.G.; Green, J.J. Polymeric nanoparticles as cancer-specific DNA delivery vectors to human hepatocellular carcinoma. J. Control. Release 2017, 263, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Botto, C.; Augello, G.; Amore, E.; Emma, M.R.; Azzolina, A.; Cavallaro, G.; Cervello, M.; Bondi, M.L. Cationic Solid Lipid Nanoparticles as Non-Viral Vectors for the Inhibition of Hepatocellular Carcinoma Growth by RNA Interference. J. Biomed. Nanotechnol. 2018, 14, 1009–1016. [Google Scholar] [CrossRef]
- Wang, D.; Chang, R.; Wang, G.; Hu, B.; Qiang, Y.; Chen, Z. Polo-like Kinase 1-targeting Chitosan Nanoparticles Suppress the Progression of Hepatocellular Carcinoma. Anticancer Agents Med. Chem. 2017, 17, 948–954. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Fernandes, A.R.; Baptista, P.V. Gold Nanoparticle Approach to the Selective Delivery of Gene Silencing in Cancer-The Case for Combined Delivery? Genes (Basel) 2017, 8, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asokan, A. CRISPR genome editing in stem cells turns to gold. Nat. Mater. 2019, 18, 1038–1039. [Google Scholar] [CrossRef]
- Taghizadeh, S.; Alimardani, V.; Roudbali, P.L.; Ghasemi, Y.; Kaviani, E. Gold nanoparticles application in liver cancer. Photodiagnosis Photodyn. Ther. 2019, 25, 389–400. [Google Scholar] [CrossRef]
- Yang, N.; Li, S.; Li, G.; Zhang, S.; Tang, X.; Ni, S.; Jian, X.; Xu, C.; Zhu, J.; Lu, M. The role of extracellular vesicles in mediating progression, metastasis and potential treatment of hepatocellular carcinoma. Oncotarget 2017, 8, 3683–3695. [Google Scholar] [CrossRef] [Green Version]
- Rinaldi, L.; Folliero, V.; Palomba, L.; Zannella, C.; Isticato, R.; Di Francia, R.; Berretta, M.; de Sio, I.; Adinolfi, L.E.; Morelli, G.; et al. Sonoporation by microbubbles as gene therapy approach against liver cancer. Oncotarget 2018, 9, 32182–32190. [Google Scholar]
- Tomizawa, M.; Shinozaki, F.; Motoyoshi, Y.; Sugiyama, T.; Yamamoto, S.; Ishige, N. Suppression of hepatocellular carcinoma cell proliferation by short hairpin RNA of frizzled 2 with Sonazoid-enhanced irradiation. Int. J. Oncol. 2016, 48, 123–129. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, Y.; Shimada, M.; Tanaka, S.; Okamamoto, M.; Miyazaki, J.; Sugimachi, K. Electroporation-mediated tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)/Apo2L gene therapy for hepatocellular carcinoma. Hum. Gene Ther. 2002, 13, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Shimada, M.; Okano, S.; Suehiro, T.; Soejima, Y.; Tomita, Y.; Maehara, Y. IL-12 gene therapy is an effective therapeutic strategy for hepatocellular carcinoma in immunosuppressed mice. J. Immunol. 2004, 173, 6635–6644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, Y.; Shimada, M.; Minagawa, R.; Tsujita, E.; Harimoto, N.; Tanaka, S.; Shirabe, K.; Miyazaki, J.; Maehara, Y. Muscle-targeted interleukin-12 gene therapy of orthotopic hepatocellular carcinoma in mice using in vivo electrosonoporation. Mol. Cancer Ther. 2004, 3, 1177–1182. [Google Scholar] [PubMed]
- Tan, A.T.; Yang, N.; Lee Krishnamoorthy, T.; Oei, V.; Chua, A.; Zhao, X.; Tan, H.S.; Chia, A.; Le Bert, N.; Low, D.; et al. Use of Expression Profiles of HBV-DNA Integrated Into Genomes of Hepatocellular Carcinoma Cells to Select T Cells for Immunotherapy. Gastroenterology 2019, 156, 1862–1876. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Qin, W.; Liu, T.; Jiang, D.; Cui, L.; Liu, X.; Fang, Y.; Tang, X.; Jin, H.; Qian, Q. PiggyBac-engineered T cells expressing a glypican-3-specific chimeric antigen receptor show potent activities against hepatocellular carcinoma. Immunobiology 2019. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.C.; Chak, C.P.; Lee, S.F.; Lai, J.M.; Zhu, X.M.; Wang, Y.X.; Sham, K.W.; Wong, C.H.; Cheng, C.H. Increased efficacies in magnetofection and gene delivery to hepatocellular carcinoma cells with ternary organic-inorganic hybrid nanocomposites. Chem. Asian J. 2013, 8, 1760–1764. [Google Scholar] [CrossRef]
- Liu, F.; Song, Y.; Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999, 6, 1258–1266. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Budker, V.; Wolff, J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999, 10, 1735–1737. [Google Scholar] [CrossRef]
- Kamimura, K.; Suda, T.; Xu, W.; Zhang, G.; Liu, D. Image-guided, lobe-specific hydrodynamic gene delivery to swine liver. Mol. Ther. 2009, 17, 491–499. [Google Scholar] [CrossRef]
- Kamimura, K.; Kanefuji, T.; Yokoo, T.; Abe, H.; Suda, T.; Kobayashi, Y.; Zhang, G.; Aoyagi, Y.; Liu, D. Safety assessment of liver-targeted hydrodynamic gene delivery in dogs. PLoS ONE 2014, 9, e107203. [Google Scholar] [CrossRef]
- Abe, H.; Kamimura, K.; Kobayashi, Y.; Ohtsuka, M.; Miura, H.; Ohashi, R.; Yokoo, T.; Kanefuji, T.; Suda, T.; Tsuchida, M.; et al. Effective Prevention of Liver Fibrosis by Liver-targeted Hydrodynamic Gene Delivery of Matrix Metalloproteinase-13 in a Rat Liver Fibrosis Model. Mol. Ther. Nucleic Acids 2016, 5, e276. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kamimura, K.; Abe, H.; Yokoo, T.; Ogawa, K.; Shinagawa-Kobayashi, Y.; Goto, R.; Inoue, R.; Ohtsuka, M.; Miura, H.; et al. Effects of Fibrotic Tissue on Liver-targeted Hydrodynamic Gene Delivery. Mol. Ther. Nucleic Acids 2016, 5, e359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cigliano, A.; Wang, C.; Pilo, M.G.; Szydlowska, M.; Brozzetti, S.; Latte, G.; Pes, G.M.; Pascale, R.M.; Seddaiu, M.A.; Vidili, G.; et al. Inhibition of HSF1 suppresses the growth of hepatocarcinoma cell lines in vitro and AKT-driven hepatocarcinogenesis in mice. Oncotarget 2017, 8, 54149–54159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, Z.; Zhang, J.; Yuan, C.; Zhang, H.; Hou, X.; Zhang, D. Enhanced shRNA delivery by the combination of polyethylenimine, ultrasound, and nanobubbles in liver cancer. Technol. Health Care 2019, 27, 263–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, L.; Hinterberger, M.; Wirnsberger, G.; Kyewski, B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 2009, 9, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Bos, R.; Marquardt, K.L.; Cheung, J.; Sherman, L.A. Functional differences between low- and high-affinity CD8(+) T cells in the tumor environment. Oncoimmunology 2012, 1, 1239–1247. [Google Scholar] [CrossRef] [Green Version]
- Abramson, J.; Giraud, M.; Benoist, C.; Mathis, D. Aire’s partners in the molecular control of immunological tolerance. Cell 2010, 140, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, S.A.; Yang, J.C.; Sherry, R.M.; Kammula, U.S.; Hughes, M.S.; Phan, G.Q.; Citrin, D.E.; Restifo, N.P.; Robbins, P.F.; Wunderlich, J.R.; et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 2011, 17, 4550–4557. [Google Scholar] [CrossRef]
- Dudley, M.E.; Yang, J.C.; Sherry, R.; Hughes, M.S.; Royal, R.; Kammula, U.; Robbins, P.F.; Huang, J.; Citrin, D.E.; Leitman, S.F.; et al. Adoptive cell therapy for patients with metastatic melanoma: Evaluation of intensive myeloablative chemoradiation preparative regimens. J. Clin. Oncol. 2008, 26, 5233–5239. [Google Scholar] [CrossRef]
- Robbins, P.F.; Morgan, R.A.; Feldman, S.A.; Yang, J.C.; Sherry, R.M.; Dudley, M.E.; Wunderlich, J.R.; Nahvi, A.V.; Helman, L.J.; Mackall, C.L.; et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011, 29, 917–924. [Google Scholar] [CrossRef]
- Sterman, D.H.; Recio, A.; Haas, A.R.; Vachani, A.; Katz, S.I.; Gillespie, C.T.; Cheng, G.; Sun, J.; Moon, E.; Pereira, L.; et al. A phase I trial of repeated intrapleural adenoviral-mediated interferon-beta gene transfer for mesothelioma and metastatic pleural effusions. Mol. Ther. 2010, 18, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Nemunaitis, J.; Barve, M.; Orr, D.; Kuhn, J.; Magee, M.; Lamont, J.; Bedell, C.; Wallraven, G.; Pappen, B.O.; Roth, A.; et al. Summary of bi-shRNA/GM-CSF augmented autologous tumor cell immunotherapy (FANG) in advanced cancer of the liver. Oncology 2014, 87, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Senzer, N.; Barve, M.; Kuhn, J.; Melnyk, A.; Beitsch, P.; Lazar, M.; Lifshitz, S.; Magee, M.; Oh, J.; Mill, S.W.; et al. Phase I trial of “bi-shRNAi(furin)/GMCSF DNA/autologous tumor cell” vaccine (FANG) in advanced cancer. Mol. Ther. 2012, 20, 679–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhanasekaran, R.; Nault, J.C.; Roberts, L.R.; Zucman-Rossi, J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology 2019, 156, 492–509. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cao, Y.; Chen, C.; Zhang, X.; McNabola, A.; Wilkie, D.; Wilhelm, S.; Lynch, M.; Carter, C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006, 66, 11851–11858. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, V.; El-Rayes, B.F.; Droz Dit Busset, M.; Cotsoglou, C.; Harris, W.P.; Damjanov, N.; Masi, G.; Rimassa, L.; Personeni, N.; Braiteh, F.; et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. Br. J. Cancer 2019, 120, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Doss, W.; Shiha, G.; Hassany, M.; Soliman, R.; Fouad, R.; Khairy, M.; Samir, W.; Hammad, R.; Kersey, K.; Jiang, D.; et al. Sofosbuvir plus ribavirin for treating Egyptian patients with hepatitis C genotype 4. J. Hepatol. 2015, 63, 581–585. [Google Scholar] [CrossRef]
- Wang, B.; Hsu, C.J.; Lee, H.L.; Chou, C.H.; Su, C.M.; Yang, S.F.; Tang, C.H. Impact of matrix metalloproteinase-11 gene polymorphisms upon the development and progression of hepatocellular carcinoma. Int. J. Med. Sci. 2018, 15, 653–658. [Google Scholar] [CrossRef] [Green Version]
- Petrini, I.; Lencioni, M.; Ricasoli, M.; Iannopollo, M.; Orlandini, C.; Oliveri, F.; Bartolozzi, C.; Ricci, S. Phase II trial of sorafenib in combination with 5-fluorouracil infusion in advanced hepatocellular carcinoma. Cancer Chemother. Pharmacol. 2012, 69, 773–780. [Google Scholar] [CrossRef]
- Majzner, R.G.; Mackall, C.L. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 2019, 25, 1341–1355. [Google Scholar] [CrossRef]
- Chen, Y.; E, C.Y.; Gong, Z.W.; Liu, S.; Wang, Z.X.; Yang, Y.S.; Zhang, X.W. Chimeric antigen receptor-engineered T-cell therapy for liver cancer. Hepatobiliary Pancreat. Dis. Int. 2018, 17, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yi, M.; Qin, S.; Wu, K. Next generation chimeric antigen receptor T cells: Safety strategies to overcome toxicity. Mol. Cancer 2019, 18, 125. [Google Scholar] [CrossRef] [PubMed]
- Ruiz de Galarreta, M.; Lujambio, A. Therapeutic editing of hepatocyte genome in vivo. J. Hepatol. 2017, 67, 818–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankowicz, F.P.; Jarrett, K.E.; Lagor, W.R.; Bissig, K.D. CRISPR/Cas9: At the cutting edge of hepatology. Gut 2017, 66, 1329–1340. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.X.; Zhang, Y.; Yin, H. Genome Editing with mRNA Encoding ZFN, TALEN, and Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Wan, T.; Xin, H.; Li, D.; Pan, H.; Wu, J.; Ping, Y. Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 2019, 21, e3107. [Google Scholar] [CrossRef] [Green Version]

| Gene Transfer Methods | Genetic Materials (Cloning Capacity)/ Functional Component | Advantages | Disadvantages | |
|---|---|---|---|---|
| Viral Vectors | ||||
| Retroviral Vectors | ||||
| Oncoretrovirus | Single stranded RNA (8 kb) | High transduction efficiency | Infect to dividing cells | Random integration Low efficiency of purification |
| Lentivirus | Single stranded RNA (8 kb) | High transduction efficiency Sustained gene expression Low immune response | Infect to dividing and non-dividing cells | Random integration Low efficiency of purification |
| Foamy virus | Single stranded RNA (9.2 kb) | High transduction efficiency Sustained gene expression No expression of viral proteins Low immune response | Infect to dividing cells Form a stable transduction intermediate in non-dividing cells | Random integration Low efficiency of purification |
| Adenoviral Vectors | ||||
| First generation adenovirus (FGAd) | Double stranded DNA (8–10 kb) | High transduction efficiency | Infect to dividing and non-dividing cells | Transient expression Host innate immune response Complicated vector production |
| Helper-dependent adenovirus (HDAd) | Double stranded DNA (~37 kb) | Large insert size Essentially no integration | Infect to dividing and non-dividing cells | Transient expression Host innate immune response Complicated vector production |
| Adeno-associated virus | Single stranded DNA (4–5 kb) | Non pathogenic Sustained gene expression Mainly no integration Low immune response | Infect to dividing and non-dividing cells | Integration may occur Small capacity of transgene Transient expression Complicated vector production |
| Herpes simplex virus | Double stranded DNA (~30 kb) | Large insert size No integration Sustained gene expression | Infectivity to nervous system | Transient expression Low transduction efficiency |
| Non-viral Vectors (Chemicals) | ||||
| Cationic lipids | Cationic charge, hydrophobic domain | High efficiency in vitro, ease to prepare | Low efficiency in vivo, acute immune response | |
| Cationic polymers | Cationic charge, polymer | Highly effective in vitro, ease to prepare | Toxic to cells, acute immune response | |
| Proteins | Natural or chemically modified proteins in cationic nature | Highly effective in vitro, less toxic, can be target specific | Low activity in vivo | |
| Peptides | Lysine or arginine residues in peptides | Highly effective in vitro, less toxic, can be target specific | Low activity in vivo | |
| Non-viral Vectors (Physical Methods) | ||||
| Needle injection | Mechanic force | Simple | Low efficiency, expression limited to needle track | |
| Gene gun | Pressure | Ease, Good efficiency | Limited to target area, need surgical procedure for internal organ | |
| Electroporation | Electric pulse | High efficiency | Tissue damage, limited target area, need surgical procedure for internal organ | |
| Sonoporation | Ultrasound | Simple, can be site-specific | Low efficiency, tissue damage | |
| Magnetofection | Magnetic field | Site specific | Low efficiency, limited target area, need surgical procedure for internal organ | |
| Hydrodynamic delivery | Hydrodynamic pressure | Simple, high efficiency in vivo, site specific | Need catheter insertion technique in large animals | |
| Immunotherapy | CAR-T, T cells | Antigen-specific | Require ex vivo cell culture Poorly effective for solid tumors | |
| Gene Vaccination | Antigen-pulsed dendritic cells | Ease to prepare less toxic, ease to administer | Low efficacy | |
| No | NCT Number | Types of Liver Tumors | Gene/Antigen | Types of Gene | Vectors or Cells | Intervention | Route of Administration | Phase | Current Status and Results | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NCT00003147 | HCC | p53 | Adenoviral Vector | Ad5CMV-p53 gene | Percutaneous injection | 1 | Terminated No results available | ||
| 2 | NCT01071941 | Primary Liver Cancers | Oncolytic Virus | Oncolytic Virus | Herpes simplex virus 1 | rRp450 | Hepatic arterial injection | 1 | Recruiting No results available Estimated Completion Date: July, 2020 | |
| Metastatic Liver Tumors | ||||||||||
| 3 | NCT00844623 | HCC | HSVtk | Suicide | Adenoviral vector | TK99UN (adenoviral vector containing TK) | Intratumoral injection | 1 | Completed Results partly reported. | [73,124] |
| 4 | NCT02202564 | HCC | HSVtk | Suicide | Adenoviral vector | LT ADV-TK ganciclovir | Intravenous infusion | 2 | Completed No results reported to date | |
| 5 | NCT02561546 | HCC | p53 | Tumor suppressor | Recombinant adenoviral vector | p53 gene therapy TAE | Hepatic arterial injection following TAE | 2 | Not yet recruiting | |
| 6 | NCT00300521 | HCC | HSVtk | Suicide | Adenoviral vector | ADV-TK | Intravenous infusion | 2 | Completed No results reported to date | |
| 7 | NCT00004178 | Primary Liver Cancers Metastatic Liver Tumors | CEA | Tumor-related Protein Coding | T Cells Modified with Chimeric Anti-CEA Immunoglobulin-T Cell Receptors (IgTCR) in Adenocarcinoma | Therapeutic autologous lymphocytes | Intravenous infusion | 1 | Completed No results reported to date | |
| 8 | NCT03313596 | HCC | HSVtk | Suicide | Adenoviral vector | ADV-TK LT | Intravenous infusion | 3 | Recruiting No results available Estimated Completion Date: Dec, 2019 | |
| 9 | NCT03480152 | Primary Liver Cancers | mRNA containing epitopes from immunogenic neoantigens | Tumor-related Protein Coding | mRNA vaccine | NCI-4650, a mRNA-based, Personalized Cancer Vaccine | Intramuscular injection | 1/2 | Terminated Related results partly reported | [159,160,161] |
| Metastatic Liver Tumors | mRNA containing epitopes from immunogenic predicted neoantigens | |||||||||
| mRNA containing epitopes from immunogenic mutations in tumor suppressor or driver genes | ||||||||||
| 10 | NCT01967823 | HCC Metastatic Liver Tumors | NY-ESO-1 | Tumor-related Protein Coding | Anti-NY ESO-1 Murine TCR-Gene Engineered Lymphocytes | Anti-NY ESO-1 mTCR PBL Cyclophosphamide Fludarabine Aldesleukin | Intravenous infusion | 2 | Recruiting No results available Estimated Completion Date: July, 2028 | [162,163,164] |
| 11 | NCT02509169 | HCC | p53 | Tumor suppressor | Recombinant adenoviral vector | TAE plus P53 gene TAE | Hepatic arterial injection following TAE | 2 | Recruiting No results available | |
| 12 | NCT02932956 | Pediatric Primary Liver Cancers | GPC-3 | Tumor-related Protein Coding | CAR T cells | GAP T cells Cytoxan Fludara | 1 | Recruiting No results available Estimated Completion Date: Feb, 2037 | ||
| 13 | NCT00012155 | Metastatic Liver Tumors | Oncoytic Virus | Oncoytic Virus | oncolytic herpes simplex virus type-1(HSV-1) | NV1020, oncolytic herpes simplex virus type-1 (HSV-1) | Hepatic arterial injection | 1 | Completed Results partly reported | [74] |
| 14 | NCT00066404 | Metastatic Liver Tumors | Interferon-beta | Genetic Immunotherapy | Adenoviral vector | recombinant adenovirus-hIFN-beta | Intrapleural injection | 1 | Active, not recruiting Results partly reported | [165] |
| 15 | NCT00035919 | Metastatic Liver Tumors | Dominant Negative Cyclin G1 | Tumor suppressor | Retroviral Vector | Mx-dnG1 Retroviral Vector | Hepatic arterial infusion | 1/2 | Withdrawn | |
| 16 | NCT00005629 | Primary Liver Cancers Metastatic Liver Tumors | AFP | Tumor-related Protein Coding | AFP peptide | AFP gene hepatocellular carcinoma vaccine | Intradermal injection | 1/2 | Completed No results reported to date | |
| 17 | NCT02905188 | HCC | GPC-3 | Tumor-related Protein Coding | CAR T cells | GLYCAR T cells | Intravenous infusion | 1 | Recruiting No results available Estimated Completion Date: Oct, 2036 | |
| 18 | NCT00093548 | HCC | AFP, GM-CSF | Tumor-related Protein Coding | Adenoviral vector | Vaccination AFP plasmid DNA vaccine GM-CSF plasmid DNA hepatocellular carcinoma vaccine adjuvant | Intramuscular injection/Intradermal injection | 1/2 | Withdrawn | [92] |
| 19 | NCT01628640 | Primary Liver Cancers Metastatic Liver Tumors | Interferon-beta | Genetic Immunotherapy | Vesicular Stomatitis Virus | Recombinant Vesicular Stomatitis Virus-expressing Interferon-beta | Intratumoral Injection | 1 | Active, not recruiting Estimated Completion Date: June, 2025 | |
| 20 | NCT03602079 | HCC CCC Metastatic Liver Tumors | HER-2 | Tumor-related Protein Coding | Antibody Drug Conjugate (ADC) | A166, an Antibody Drug Conjugate (ADC) targeting HER2 expressing cancer cells. | Intravenous infusion | 1/2 | Recruiting No results available Estimated Completion Date: May, 2021 | |
| 21 | NCT02416466 | Metastatic Liver Tumors | CEA | Tumor-related Protein Coding | CAR-T cells | anti-CEA CAR-T cells | Hepatic arterial infusion | 1 | Completed No results reported to date | |
| 22 | NCT02869217 | NY-ESO-1 Expressing Liver Cancers in HLA-A2 Positive Patients Metastatic Liver Tumors | NY-ESO-1 | Tumor-related Protein Coding | NY-ESO-1 Specific TCR Gene Transduced Autologous T Lymphocytes | TBI-1301 (NY-ESO-1 Specific TCR Gene Transduced Autologous T Lymphocytes) Cyclophosphamide | Infusion | 1 | Recruiting No results available Estimated Completion Date: June, 2020 | |
| 23 | NCT01061840 | Primary Liver Cancers Metastatic Liver Tumors | rhGMCSF and bi-shRNAfurin from the Vigil™ plasmid | Tumor-related Protein Coding | plasmid | Vaccination | Intradermal injection | 1 | Completed Results partly reported | [166,167] |
| 24 | NCT01437007 | Metastatic Liver Tumors from Colorectal, Pancreas, Gastric, Breast, and Ovarian Cancers | siRNA Against the PLK1 | Oncogene suppression | Lipid Nanoparticles | TKM-080301 | Hepatic arterial infusion | 1 | Completed Results partly reported | [53] |
| 25 | NCT03971747 | HCC | AFP | Tumor-related Protein Coding | T Cell | AFP Specific T Cell Receptor T Cells | Intravenous infusion | 1 | Not yet recruiting | |
| 26 | NCT02418988 | HCC | p53 | Tumor suppressor | Recombinant adenoviral vector | TACE plus rAd-p53 artery injection TACE | Injected into the embolization artery. | 2 | Recruiting No results available | |
| 27 | NCT02850536 | Metastatic Liver Tumors from Colorectal, Pancreas, Gastric, Breast, and Ovarian Cancers | CEA | Tumor-related Protein Coding | CAR-T cells | anti-CEA CAR-T cells | Hepatic arterial infusion or splenic vein | 1 | Active, not recruiting Estimated Completion Date: Dec, 2019 | |
| 28 | NCT01373047 | Metastatic Liver Tumors from Colorectal, Pancreas, Gastric, Breast, and Ovarian Cancers | CEA | Tumor-related Protein Coding | T cell | anti-CEA 2nd generation designer T cells | Hepatic arterial infusion or splenic vein | 1 | Completed No results reported to date | |
| 29 | NCT02432963 | Adult Solid Neoplasm | p53 | Tumor suppressor | Modified vaccinia virus | Vaccination | 1 | Active, not recruiting Estimated Completion Date: Feb, 2020 | ||
| 30 | NCT02715362 | HCC | GPC3 | Tumor-related Protein Coding | CAR-T cells | TAI-GPC3-CART cells | Hepatic arterial infusion | 1/2 | Recruiting No results available | |
| 31 | NCT00301106 | Metastatic Liver Tumors from Colorectal, Pancreas, Gastric, Breast, and Ovarian Cancers | Interleukin-12 | Genetic Immunotherapy | Adenoviral Vector | adenovirus-mediated human interleukin-12 | Intratumoral Injection | 1 | Terminated No results available | |
| 32 | NCT00072098 | Metastatic Liver Tumors from Colorectal, Pancreas, Gastric, Breast, and Ovarian Cancers | Interleukin-12 | Genetic Immunotherapy | Adenoviral Vector | adenoviral vector-delivered interleukin-12 | Intratumoral Injection | 1 | Terminated No results available | |
| 33 | NCT03198546 | HCC | GPC3 | Tumor-related Protein Coding | CAR-T cells | GPC3 targeting CAR-T cells | Systemic or local injections | 1 | Recruiting No results available Estimated Completion Date: Aug, 2022 | [87] |
| 34 | NCT00103142 | Metastatic Liver Tumors from Colorectal, Pancreas, Gastric, Breast, and Ovarian Cancers | Vaccinia-Carcinoembryonic antigen (CEA)-mucin 1 (MUC-1)- Triad of costimulatory molecules TRICOM vaccine (PANVAC-V) | Tumor-related Protein Coding | Autologous dendritic cells | Vaccination | 2 | Completed Results available | [95] |
| No | NCT Number | Official Title | Brief Summary | Types of Liver Tumors | Phase | Enrollment | Current Status and Results | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | NCT00373737 | Microarray Analysis of Gene Expression in Liver Tumors | This study aims to study the gene expression profiles of liver tumors to help us understand their biology, and to find new tumor and treatment markers for liver cancer. | Liver Cancer | Not Applicable | 300 | Completed No results reported to date | |
| 2 | NCT01786980 | The Methylation Phenotype Screening and Determination Mode Study of Liver Cancer Prognosis Related Gene | This study aimed to obtain the important factors affecting liver cancer prognosis, survival, recurrence and metastasis in order to be able to find and establish the effective prognostic evaluation method by analyzing clinical information combining the information of gene chip, methylation chip and flow cytometry to carry out comprehensive researches on liver cancer cell genetics, epigenetics, stem cells and tumor microenvironment changes. | HCC | Not Applicable | 300 | Completed No results reported to date | |
| 3 | NCT00160940 | Differential Gene Expression in Liver Tissue and Blood From Individuals With Chronic Viral Hepatitis With or Without a Complicating Hepatoma or Autoimmune Liver Disease | This study aimed to find the genes that are expressed in both the circulating white blood cells and the liver of patients, using differential gene expression analysis, with varying degrees of liver damage of different causes with or without liver cancers. | Primary Liver Cancers Metastatic Liver Tumors | Not Applicable | 200 | Recruiting No results available | |
| 4 | NCT01643499 | A Genotype-guided Dosing Study of mFOLFIRINOX in Previously Untreated Patients With Advanced Gastrointestinal Malignancies | This study aimed to determine the dose of a chemotherapy drug (irinotecan) in 1st cycle in each of two UGT1A1 genotype groups (*1*1, *1*28) using genotype-guided dosing, that can be tolerated as part of a combination of drugs. | HCC CCC Metastatic Liver Tumors | 1 | 79 | Active, not recruiting Estimated Completion Date: Apr, 2019 | |
| 5 | NCT02465060 | Molecular Analysis for Therapy Choice (MATCH) | This phase II MATCH trial aimed to study how well treatment that is directed by genetic testing works in patients with solid tumors or lymphomas that have progressed following at least one line of standard treatment or for which no agreed upon treatment approach exists. | Primary Liver Cancers Metastatic Liver Tumors | 2 | 6452 | Recruiting No results available Estimated Primary Completion Date: June, 2022 | |
| 6 | NCT02733809 | Mechanism of Sorafenib Resistance in Patients With Advanced Hepatocellular Carcinoma | This study aimed to clarify the hypothesis that resistant tumor may be due to genetic mutations and/or other alternative pathways that could be the reason to overcome the SOR and still proliferate by analyzing the gene expression profiling signature (a set of dysregulated genes) for molecular classification, diagnosis, and prognosis of several types of cancers. | HCC | 4 | 40 | Recruiting No results available Estimated Completion Date: Dec, 2024 | [169] |
| 7 | NCT01752920 | A Phase 1/2 Study of ARQ 087 in Adult Subjects With Advanced Solid Tumors With FGFR Genetic Alterations, Including Intrahepatic Cholangiocarcinoma With FGFR2 Gene Fusion | This open-label, Phase 1/2, dose escalation and signal finding study aimed to clarify the effect of Derazantinib (ARQ 087), multi-kinase inhibitor designed to preferentially inhibit the FGFR family of kinases, in the cases with cholangiocarcinoma with FGFR2 gene alterations. | CCC | 1/2 | 119 | Completed Results partly reported | [170] |
| 8 | NCT03993873 | A Phase 1, Open-Label, Multi-Center, First-in-Human Study of the Safety, Tolerability, Pharmacokinetics, and Anti-Tumor Activity of TPX-0022, a Novel MET/CSF1R/SRC Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in MET | A phase 1, first-in-human, open-label study to determine the safety, tolerability, PK, and preliminary efficacy of the novel MET/CSF1R/SRC inhibitor TPX-0022 in adult subjects with advanced solid tumors harboring genetic alterations in MET. The study will proceed in three parts: a dose-escalation, a food effect, and dose-expansion. | Advanced Solid Tumor Metastatic Solid Tumors | 1 | 120 | Recruiting No results available Estimated Completion Date: Nov, 2023 | |
| 9 | NCT01892072 | VEGF Signaling Promotes Cell Growth and Metastasis in Hepatocellular Carcinoma in a VEGF Receptor Mediated Pathway | This study aimed to examine the VEGF signaling in HCC cell lines and its mechanism in HCC growth, proliferation and apoptosis. | HCC | Not Applicable | 50 | Active, not recruiting | |
| 10 | NCT02507882 | Impact of IL-28B rs12979860 and rs4803217 Gene Polymorphisms Associated With miRNAs Deregulation on HCV-related Hepatocellular Carcinoma | This study aimed to determine through investigating a cohort of 405 patients, whether IL28B rs12979860 and rs4803217 polymorphisms are associated to the risk of HCC in chronic hepatitis C patients. | HCC | Not Applicable | 405 | Not yet recruiting | |
| 11 | NCT00858000 | Analysis of the Incidence of Expression of a Specific Set of Genes and of Tumor Antigens in Cancer Tissue From Patients With Hepatocellular Carcinoma | This study aimed to analyze the expression of specific markers in HCC and tumor-related antigens to develop new approaches to treat this type of cancer with genetic immunotherapy. | HCC | Not Applicable | 30 | Completed No results reported to date | |
| 12 | NCT03722628 | The Assessment of Matrix Metalloproteinase-1 Genotypes Polymorphism as a Risk Factor for Hepatocellular Carcinoma in Chronic Hepatitis C Patients With Liver Cirrhosis | This study aimed to assess whether the Matrix Metalloproteinase-1 genotypes polymorphism can be a risk factor for HCC in chronic hepatitis C patients with liver cirrhosis. | HCC | Not Applicable | 200 | Not yet recruiting | [172] |
| 13 | NCT01930383 | Circulating Tumor Cells as Biomarkers of Prognosis and Predictors of Efficacy of Drug Therapy for Patients With Hepatocellular Carcinoma | This study aimed to explore the clinical value of correlation between circulating tumor cells numbers and other clinical characteristics in HCC patients with different stages. | HCC | Not Applicable | 150 | Recruiting No results available | |
| 14 | NCT00619541 | Phase II Study of Sorafenib (Bay 43-9006) and Infusional 5-Fluorouracil in Advanced Hepatocellular Carcinoma. | The purpose of this study is to use SOR + 5-FU to evaluate activity, efficacy, safety, pharmacodynamics and pharmacokinetics in patients with advanced HCC. | HCC | 2 | 46 | Completed Results partly reported | [173] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamimura, K.; Yokoo, T.; Abe, H.; Terai, S. Gene Therapy for Liver Cancers: Current Status from Basic to Clinics. Cancers 2019, 11, 1865. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11121865
Kamimura K, Yokoo T, Abe H, Terai S. Gene Therapy for Liver Cancers: Current Status from Basic to Clinics. Cancers. 2019; 11(12):1865. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11121865
Chicago/Turabian StyleKamimura, Kenya, Takeshi Yokoo, Hiroyuki Abe, and Shuji Terai. 2019. "Gene Therapy for Liver Cancers: Current Status from Basic to Clinics" Cancers 11, no. 12: 1865. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11121865