Establishment of an Immunocompetent Metastasis Rat Model with Hepatocyte Cancer Stem Cells
Abstract
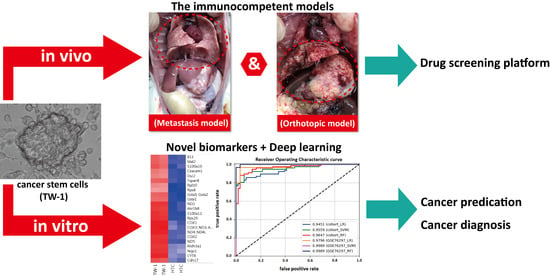
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Diethylnitrosamine (DEN)-Induced Rat Liver Cancer
2.2. Cell Culture
2.3. RNA Extraction and Microarray Assay
2.4. Western Blotting
2.5. Immunofluorescence Staining
2.6. Chemotherapy Resistance Assay
2.7. Xenograft in Nude Mice Model and the Orthotopic and Metastasis Immunocompetent Rat Model
2.8. Microarray Datasets
2.9. Biomarker Selection with Single Biomarker AUC Scores
2.10. Experimental Details for Machine Learning Classifiers
2.11. Statistical Analyses
3. Results
3.1. Isolation and Culture of Novel Liver Cancer Stem Cell Lines
3.2. TW-1 Showed the Ability to Differentiate
3.3. TW-1 Shows a High Chemotherapy Resistance In Vitro and In Vivo
3.4. TW-1 Cells Showed Metastases in Wild-Type Rats
3.5. Prediction of HCC with Machine Learning Classifiers
4. Discussion
4.1. Drug Resistance and Cancer Recurrence
4.2. Liver Cancer Stem Cells and Mitochondrial Metabolism
4.3. Metastasis Model in Immunocompetent Rats
4.4. Biomarkers of HCC in Clinical Application
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| CSCs | Cancer stem cells |
| EpCAM | epithelial cell adhesion molecule |
| DEN | Diethylnitrosamine |
| RMA | multi-array average |
| LR | logistic regression |
| SVM | support vector machine |
| RF | random forest |
| ROC | receiver operating characteristic |
| AUC | area under the ROC curve |
| AFP | alpha-fetoprotein |
| ABC | ATP-binding cassette |
| CYP | cytochrome P450 system |
| GST | glutathione-S-transferase |
| UGT | uridine diphospho-glucuronosyltransferase |
| MAPK | mitogen-activated protein kinase |
| TME | tumor microenvironment |
| OXPHOS | oxidative phosphorylation |
References
- Wen, P.H.; Lu, C.L.; Strong, C.; Lin, Y.J.; Chen, Y.L.; Li, C.Y.; Tsai, C.C. Demographic and Urbanization Disparities of Liver Transplantation in Taiwan. Int. J. Environ. Res. Public Health 2018, 15, 177. [Google Scholar] [CrossRef] [Green Version]
- Takeishi, K.; Maeda, T.; Tsujita, E.; Yamashita, Y.; Harada, N.; Itoh, S.; Harimoto, N.; Ikegami, T.; Yoshizumi, T.; Shirabe, K.; et al. Predictors of intrahepatic multiple recurrences after curative hepatectomy for hepatocellular carcinoma. Anticancer Res. 2015, 35, 3061–3066. [Google Scholar]
- Dick, J.E. Breast cancer stem cells revealed. Proc. Natl. Acad. Sci. USA 2003, 100, 3547–3549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paula, A.D.C.; Lopes, C. Implications of Different Cancer Stem Cell Phenotypes in Breast Cancer. Anticancer Res. 2017, 37, 2173–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, L.A.; Lopez-Gil, J.C.; Sainz, B., Jr.; Hermann, P.C. The Cancer Stem Cell in Hepatocellular Carcinoma. Cancers 2020, 12, 684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.T.; Ryu, C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017, 50, 285–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S. Biology and clinical implications of CD133+ liver cancer stem cells. Exp. Cell Res. 2013, 319, 126–132. [Google Scholar] [CrossRef]
- Slomiany, M.G.; Dai, L.; Tolliver, L.B.; Grass, G.D.; Zeng, Y.; Toole, B.P. Inhibition of Functional Hyaluronan-CD44 Interactions in CD133-positive Primary Human Ovarian Carcinoma Cells by Small Hyaluronan Oligosaccharides. Clin. Cancer Res. 2009, 15, 7593–7601. [Google Scholar] [CrossRef] [Green Version]
- Wahab, S.M.R.; Islam, F.; Gopalan, V.; Lam, A.K.-Y. The Identifications and Clinical Implications of Cancer Stem Cells in Colorectal Cancer. Clin. Colorectal Cancer 2017, 16, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zuo, X.; Xie, K.; Wei, D. The Role of CD44 and Cancer Stem Cells. Bioinform. MicroRNA Res. 2017, 1692, 31–42. [Google Scholar] [CrossRef]
- Galuppo, R.; Maynard, E.; Shah, M.; Daily, M.F.; Chen, C.; Spear, B.T.; Gedaly, R. Synergistic inhibition of HCC and liver cancer stem cell proliferation by targeting RAS/RAF/MAPK and WNT/beta-catenin pathways. Anticancer Res. 2014, 34, 1709–1713. [Google Scholar] [PubMed]
- Pu, H.; Zheng, Q.; Li, H.; Wu, M.; Tianming, L.; Gui, X.; Li, T.; Lu, D. CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget 2015, 6, 40775–40798. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ji, J.; Xu, Y.; Liu, Y.; Shi, L.; Liu, Y.; Lu, X.; Zhao, Y.; Luo, F.; Wang, B.; et al. Faculty Opinions recommendation of MicroRNA-191, by promoting the EMT and increasing CSC-like properties, is involved in neoplastic and metastatic properties of transformed human bronchial epithelial cells. Mol. Carcinog. 2017, 54 (Suppl. 1), 148–161. [Google Scholar] [CrossRef]
- Yamashita, T.; Wang, X.W. Cancer stem cells in the development of liver cancer. J. Clin. Investig. 2013, 123, 1911–1918. [Google Scholar] [CrossRef]
- Caviglia, J.M.; Eferl, R. Mouse Models of Liver Cancer. Bioinform. MicroRNA Res. 2015, 1267, 165–183. [Google Scholar] [CrossRef]
- Huang, K.-F.; Hsu, W.-C.; Hsiao, J.-K.; Chen, G.-S.; Wang, J.-Y. Collagen-Glycosaminoglycan Matrix Implantation Promotes Angiogenesis following Surgical Brain Trauma. BioMed. Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Chen, H.-L.; Chien, C.-S.; Wu, S.-H.; Ho, Y.-T.; Yu, C.-H.; Chang, M.-H. Contribution of Mature Hepatocytes to Biliary Regeneration in Rats with Acute and Chronic Biliary Injury. PLoS ONE 2015, 10, e0134327. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.-S.; Chen, Y.-H.; Chen, H.-L.; Wang, C.-P.; Wu, S.-H.; Ho, S.-L.; Huang, W.-C.; Yu, C.-H.; Chang, M.-H. Cells responsible for liver mass regeneration in rats with 2-acetylaminofluorene/partial hepatectomy injury. J. Biomed. Sci. 2018, 25, 39. [Google Scholar] [CrossRef]
- Yu, Z.; Pestell, T.G.; Lisanti, M.P.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef] [Green Version]
- Furey, T.S.; Cristianini, N.; Duffy, N.; Bednarski, D.W.; Schummer, M.; Haussler, D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 2000, 16, 906–914. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.-B.; Li, Y.-M.; Zhao, Y.; Wang, R.; Wu, Q.; Zheng, R.-S.; Ou, Y.-R. Role of IL-33 expression in oncogenesis and development of human hepatocellular carcinoma. Oncol. Lett. 2016, 12, 429–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Liu, X.-K.; Chu, Z.; Ye, J.C.; Li, K.-L.; Zhuang, W.-L.; Yang, D.-J.; Jiang, Y.-F. Detection of Interleukin-33 in Serum and Carcinoma Tissue from Patients with Hepatocellular Carcinoma and its Clinical Implications. J. Int. Med. Res. 2012, 40, 1654–1661. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Li, H.; Yu, Y.; Chen, J.; Chen, X.; Ren, F.; Ren, Z.; Cui, G. Enolase-1 serves as a biomarker of diagnosis and prognosis in hepatocellular carcinoma patients. Cancer Manag. Res. 2018, 10, 5735–5745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S.; Housman, G.; Byler, S.; Heerboth, S.; et al. Drug Resistance in Cancer: An Overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [Green Version]
- Michael, M.; Doherty, M. Tumoral Drug Metabolism: Overview and Its Implications for Cancer Therapy. J. Clin. Oncol. 2005, 23, 205–229. [Google Scholar] [CrossRef]
- Rodriguezantona, C.; Ingelman-Sundberg, M. Cytochrome P450 pharmacogenetics and cancer. Oncogene 2006, 25, 1679–1691. [Google Scholar] [CrossRef] [Green Version]
- Townsend, D.M.; Tew, K.D. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 2003, 22, 7369–7375. [Google Scholar] [CrossRef] [Green Version]
- Cummings, J.; Ethell, B.T.; Jardine, L.; Boyd, G.; MacPherson, J.S.; Burchell, B.; Smyth, J.F.; Jodrell, D.I. Glucuronidation as a mechanism of intrinsic drug resistance in human colon cancer: Reversal of resistance by food additives. Cancer Res. 2003, 63, 8443–8450. [Google Scholar]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Song, I.S.; Jeong, J.Y.; Jeong, S.H.; Kim, H.K.; Ko, K.S.; Rhee, B.D.; Kim, N.; Han, J. Mitochondria as therapeutic targets for cancer stem cells. World J. Stem Cells 2015, 7, 418–427. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The Cancer Stem Cell Niche: How Essential Is the Niche in Regulating Stemness of Tumor Cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Mu, Q.; Huang, H. The Roles of Insulin-Like Growth Factor 2 mRNA-Binding Protein 2 in Cancer and Cancer Stem Cells. Stem Cells Int. 2018, 2018, 4217259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, Y.H.; Yook, J.I.; Kim, H.S.; Kim, N.H. Catabolic metabolism during cancer EMT. Arch. Pharmacal Res. 2015, 38, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Park, J.H.; Jung, K.H.; Levine, H.; Kaipparettu, B.A. Elucidating the Metabolic Plasticity of Cancer: Mitochondrial Reprogramming and Hybrid Metabolic States. Cells 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, M.-K.; Cheong, J.-H. Mitochondria-centric bioenergetic characteristics in cancer stem-like cells. Arch. Pharmacal Res. 2019, 42, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Wolpaw, A.J.; Dang, C.V. Exploiting Metabolic Vulnerabilities of Cancer with Precision and Accuracy. Trends Cell Biol. 2018, 28, 201–212. [Google Scholar] [CrossRef]
- Tesori, V.; Piscaglia, A.C.; Samengo, D.; Barba, M.; Bernardini, C.; Scatena, R.; Pontoglio, A.; Castellini, L.; Spelbrink, J.N.; Maulucci, G.; et al. The multikinase inhibitor Sorafenib enhances glycolysis and synergizes with glycolysis blockade for cancer cell killing. Sci. Rep. 2015, 5, srep09149. [Google Scholar] [CrossRef] [Green Version]
- Wolf, D.A. Is Reliance on Mitochondrial Respiration a “Chink in the Armor” of Therapy-Resistant Cancer? Cancer Cell 2014, 26, 788–795. [Google Scholar] [CrossRef] [Green Version]
- Afify, S.M.; Hassan, G.; Osman, A.; Calle, A.S.; Nawara, H.M.; Zahra, M.H.; El-Ghlaban, D.; Mansour, H.; Alam, M.J.; Abu Quora, H.A.; et al. Metastasis of Cancer Stem Cells Developed in the Microenvironment of Hepatocellular Carcinoma. Bioengineering 2019, 6, 73. [Google Scholar] [CrossRef] [Green Version]
- Leung, M.K.K.; Xiong, H.Y.; Lee, L.J.; Frey, B.J. Deep learning of the tissue-regulated splicing code. Bioinformatics 2014, 30, i121–i129. [Google Scholar] [CrossRef] [Green Version]
- Salakhutdinov, R.; Hinton, G. An Efficient Learning Procedure for Deep Boltzmann Machines. Neural Comput. 2012, 24, 1967–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene ID | Fold Change (TW-1/HTC) | Gene Symbol | Description |
|---|---|---|---|
| 17867292 | 718.25 | ND2 | NADH dehydrogenase subunit 2 |
| 17782314 | 613.15 | Akr1b8 | aldo-keto reductase family 1, member B8 |
| 17740581 | 586.22 | S100a11 | S100 calcium binding protein A11 |
| 17822248 | 573.16 | Rps29 | ribosomal protein S29 |
| 17880082 | 383.61 | Rpl10 | ribosomal protein L10 |
| 17867310 | 359.91 | ND4; ND4L | NADH dehydrogenase subunit 4; NADH dehydrogenase subunit 4L |
| 17867316 | 306.48 | ND5 | NADH dehydrogenase subunit 5 |
| 17740584 | 292.64 | S100a10 | S100 calcium binding protein A10 |
| 17863382 | 280.59 | Gsta1 | glutathione S-transferase alpha 1 |
| 17867302 | 252.12 | COX3; ND3; ATP6 | cytochrome c oxidase subunit 3; NADH dehydrogenase subunit 3; ATPase subunit 6 |
| 17867294 | 248.4 | COX1 | cytochrome c oxidase subunit 1 |
| 17846131 | 244.88 | Gsta5; Gsta2 | glutathione S-transferase alpha 5; glutathione S-transferase alpha 2 |
| 17809643 | 231.16 | Rps8 | ribosomal protein S8 |
| 17867298 | 229.75 | COX2 | COXII |
| 17624945 | 165.35 | Il33 | interleukin 33 |
| 17829386 | 153.32 | Mal2 | mal, T-cell differentiation protein 2 |
| 17867318 | 150.73 | CYTB | cytochrome b |
| 17646585 | 146.19 | Aldh3a1 | aldehyde dehydrogenase 3 family, member A1 |
| 17661655 | 132.48 | Coa3 | cytochrome C oxidase assembly factor 3 |
| 17630680 | 131.89 | Ceacam1 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| 17828240 | 125.69 | Tspan8 | tetraspanin 8 |
| 17683922 | 120.28 | Cd55 | CD55 molecule, decay accelerating factor for complement |
| 17733363 | 118.79 | Nqo1 | NAD(P)H dehydrogenase, quinone 1 |
| 17832979 | 112.26 | Rpl41 | ribosomal protein L41 |
| 17715346 | 108.73 | Serpinb6 | serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| 17721215 | 106.48 | Prtfdc1 | phosphoribosyl transferase domain containing 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Tseng, I.-C.; Huang, W.-C.; Su, C.-W.; Lai, Y.-H.; Lin, C.; Lee, A.Y.-L.; Kuo, C.-Y.; Su, L.-Y.; Lee, M.-C.; et al. Establishment of an Immunocompetent Metastasis Rat Model with Hepatocyte Cancer Stem Cells. Cancers 2020, 12, 3721. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12123721
Wu S, Tseng I-C, Huang W-C, Su C-W, Lai Y-H, Lin C, Lee AY-L, Kuo C-Y, Su L-Y, Lee M-C, et al. Establishment of an Immunocompetent Metastasis Rat Model with Hepatocyte Cancer Stem Cells. Cancers. 2020; 12(12):3721. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12123721
Chicago/Turabian StyleWu, Semon, I-Chieh Tseng, Wen-Cheng Huang, Cheng-Wen Su, Yu-Heng Lai, Che Lin, Alan Yueh-Luen Lee, Chan-Yen Kuo, Li-Yu Su, Ming-Cheng Lee, and et al. 2020. "Establishment of an Immunocompetent Metastasis Rat Model with Hepatocyte Cancer Stem Cells" Cancers 12, no. 12: 3721. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12123721