Fully Human Antibodies for Malignant Pleural Mesothelioma Targeting
Abstract
:1. Introduction
2. Immunotherapy in MPM
3. Making a Hot Tumor Microenvironment
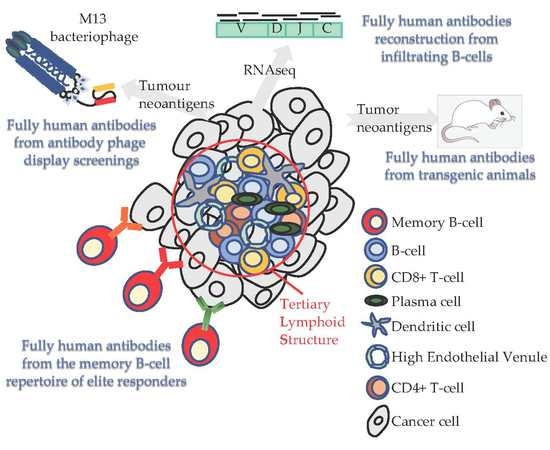
4. Tertiary Lymphoid Structures in Solid Tumors and MPM: Where the Anti-Tumor Response Begins
5. The Importance of B-cell Infiltration in Solid Tumors and MPM
6. The Quest for Specificity in Malignant Mesothelioma: How Do We Fill This Gap?
7. Phage Display Screening Using Human Synthetic Antibody Libraries
8. Fully-Human Antibodies from Genetically Modified Transgenic Mice
9. From Today’s Patients the Future Cures for MPM
10. BCR Repertoire from Sequencing Data
11. MPM Patient Memory B-cell Receptor Repertoire Exploitation to Find Novel Therapeutic Antibodies
12. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carbone, M.; Adusumilli, P.S.; Alexander, H.R., Jr.; Baas, P.; Bardelli, F.; Bononi, A.; Bueno, R.; Felley Bosco, E.; Galateau Salle, F.; Jablons, D.; et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J. Clin. 2019, 69, 402–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kameda, T.; Takahashi, K.; Kim, R.; Jiang, Y.; Movahed, M.; Park, E.-K.; Rantanen, J. Asbestos: Use, bans and disease burden in Europe. Bull. World Health Organ. 2014, 92, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Peto, J.; Decarli, A.; La Vecchia, C.; Levi, F.; Negri, E. The European mesothelioma epidemic. Br. J. Cancer 1999, 79, 666–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henley, S.J.; Larson, T.C.; Wu, M.; Antao, V.C.S.; Lewis, M.; Pinheiro, G.A.; Eheman, C. Mesothelioma incidence in 50 states and the District of Columbia, United States, 2003–2008. Int. J. Occup. Environ. Health 2013, 19, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckett, P.; Edwards, J.; Fennell, D.; Hubbard, R.; Woolhouse, I.; Peake, M.D. Demographics, management and survival of patients with malignant pleural mesothelioma in the National Lung Cancer Audit in England and Wales. Lung Cancer 2015, 88, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.; Testa, J.R. BAP1, a tumor suppressor gene driving malignant mesothelioma. Transl. Lung Cancer Res. 2017, 6, 270–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Pak, H.; Hammond-Martel, I.; Ghram, M.; Rodrigue, A.; Daou, S.; Barbour, H.; Corbeil, L.; Hébert, J.; Drobetsky, E.; et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2014, 111, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Carbone, M.; Yang, H.; Pass, H.I.; Krausz, T.; Testa, J.R.; Gaudino, G. BAP1 and cancer. Nat. Publ. Group 2013, 13, 153–159. [Google Scholar] [CrossRef]
- Affar, E.B.; Carbone, M. BAP1 regulates different mechanisms of cell death. Cell Death Dis. 2018, 9, 1151–1153. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, J.; Liu, X.; Feng, L.; Gong, Z.; Koppula, P.; Sirohi, K.; Li, X.; Wei, Y.; Lee, H.; et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 2018, 20, 1181–1192. [Google Scholar] [CrossRef]
- Dey, A.; Seshasayee, D.; Noubade, R.; French, D.M.; Liu, J.; Chaurushiya, M.S.; Kirkpatrick, D.S.; Pham, V.C.; Lill, J.R.; Bakalarski, C.E.; et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 2012, 337, 1541–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bononi, A.; Giorgi, C.; Patergnani, S.; Larson, D.; Verbruggen, K.; Tanji, M.; Pellegrini, L.; Signorato, V.; Olivetto, F.; Pastorino, S.; et al. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature 2017, 546, 549–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bononi, A.; Yang, H.; Giorgi, C.; Patergnani, S.; Pellegrini, L.; Su, M.; Xie, G.; Signorato, V.; Pastorino, S.; Morris, P.; et al. Germline BAP1 mutations induce a Warburg effect. Cell Death Differ. 2017, 24, 1694–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betti, M.; Casalone, E.; Ferrante, D.; Aspesi, A.; Morleo, G.; Biasi, A.; Sculco, M.; Mancuso, G.; Guarrera, S.; Righi, L.; et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017, 405, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Morrow, B.; Thomas, A.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Gadiraju, M.; Panou, V.; Gao, S.; Mian, I.; et al. Inherited predisposition to malignant mesothelioma and overall survival following platinum chemotherapy. Proc. Natl. Acad. Sci. USA 2019, 116, 9008–9013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farzin, M.; Toon, C.W.; Clarkson, A.; Sioson, L.; Watson, N.; Andrici, J.; Gill, A.J. Loss of expression of BAP1 predicts longer survival in mesothelioma. Pathology 2015, 47, 302–307. [Google Scholar] [CrossRef]
- Srinivasan, G.; Sidhu, G.S.; Williamson, E.A.; Jaiswal, A.S.; Najmunnisa, N.; Wilcoxen, K.; Jones, D.; George, T.J.; Hromas, R. Synthetic lethality in malignant pleural mesothelioma with PARP1 inhibition. Cancer Chemother. Pharm. 2017, 80, 861–867. [Google Scholar] [CrossRef] [Green Version]
- Zauderer, M.G.; Szlosarek, P.; Le Moulec, S.; Popat, S.; Taylor, P.; Planchard, D.; Scherpereel, A.; Jahan, T.; Koczywas, M.; Forster, M.; et al. Phase 2, multicenter study of the EZH2 inhibitor tazemetostat as monotherapy in adults with relapsed or refractory (R/R) malignant mesothelioma (MM) with BAP1 inactivation. JCO 2018, 36, 8515-8515. [Google Scholar] [CrossRef]
- Nicolini, F.; Bocchini, M.; Bronte, G.; Delmonte, A.; Guidoboni, M.; Crinò, L.; Mazza, M. Malignant Pleural Mesothelioma: State-of-the-Art on Current Therapies and Promises for the Future. Front. Oncol. 2019, 9, 1519. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasello, G.; Zago, G.; Lunardi, F.; Urso, L.; Kern, I.; Vlacic, G.; Grosso, F.; Mencoboni, M.; Ceresoli, G.L.; Schiavon, M.; et al. Malignant pleural mesothelioma immune microenvironment and checkpoint expression: Correlation with clinical-pathological features and intratumor heterogeneity over time. Ann. Oncol. 2018, 29, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Marcq, E.; Siozopoulou, V.; De Waele, J.; van Audenaerde, J.; Zwaenepoel, K.; Santermans, E.; Hens, N.; Pauwels, P.; van Meerbeeck, J.P.; Smits, E.L.J. Prognostic and predictive aspects of the tumor immune microenvironment and immune checkpoints in malignant pleural mesothelioma. Oncoimmunology 2017, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Nowak, A.K.; Forde, P.M. Immunotherapy trials in mesothelioma - promising results, but don’t stop here. Nat. Rev. Clin. Oncol. 2019, 16, 726–728. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [Green Version]
- Chapel, D.B.; Stewart, R.; Furtado, L.V.; Husain, A.N.; Krausz, T.; Deftereos, G. Tumor PD-L1 expression in malignant pleural and peritoneal mesothelioma by Dako PD-L1 22C3 pharmDx and Dako PD-L1 28-8 pharmDx assays. Hum. Pathol. 2019, 87, 11–17. [Google Scholar] [CrossRef]
- Brosseau, S.; Danel, C.; Scherpereel, A.; Mazieres, J.; Lantuejoul, S.; Margery, J.; Greillier, L.; Audigier-Valette, C.; Gounant, V.; Antoine, M.; et al. Shorter Survival in Malignant Pleural Mesothelioma Patients With High PD-L1 Expression Associated With Sarcomatoid or Biphasic Histology Subtype: A Series of 214 Cases From the Bio-MAPS Cohort. Clin. Lung Cancer 2019, 20, e564–e575. [Google Scholar] [CrossRef]
- Nguyen, B.H.; Montgomery, R.; Fadia, M.; Wang, J.; Ali, S. PD-L1 expression associated with worse survival outcome in malignant pleural mesothelioma. Asia Pac. J. Clin. Oncol. 2018, 14, 69–73. [Google Scholar] [CrossRef]
- Marcq, E.; Waele, J.D.; Audenaerde, J.V.; Lion, E.; Santermans, E.; Hens, N.; Pauwels, P.; van Meerbeeck, J.P.; Smits, E.L.J. Abundant expression of TIM-3, LAG-3, PD-1 and PD-L1 as immunotherapy checkpoint targets in effusions of mesothelioma patients. Oncotarget 2017, 8, 89722–89735. [Google Scholar] [CrossRef] [Green Version]
- Cedrés, S.; Ponce-Aix, S.; Zugazagoitia, J.; Sansano, I.; Enguita, A.; Navarro-Mendivil, A.; Martinez-Marti, A.; Martinez, P.; Felip, E. Analysis of expression of programmed cell death 1 ligand 1 (PD-L1) in malignant pleural mesothelioma (MPM). PLoS ONE 2015, 10, e0121071. [Google Scholar]
- Gennen, K.; Käsmann, L.; Taugner, J.; Eze, C.; Karin, M.; Roengvoraphoj, O.; Neumann, J.; Tufman, A.; Orth, M.; Reu, S.; et al. Prognostic value of PD-L1 expression on tumor cells combined with CD8+ TIL density in patients with locally advanced non-small cell lung cancer treated with concurrent chemoradiotherapy. Radiat. Oncol. 2020, 15, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhao, B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: Meta-analysis. BMJ 2018, 362, k3529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conroy, J.M.; Pabla, S.; Nesline, M.K.; Glenn, S.T.; Papanicolau-Sengos, A.; Burgher, B.; Andreas, J.; Giamo, V.; Wang, Y.; Lenzo, F.L.; et al. Next generation sequencing of PD-L1 for predicting response to immune checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Lantuejoul, S.; Le Stang, N.; Damiola, F.; Scherpereel, A.; Galateau Salle, F. PD-L1 Testing for Immune Checkpoint Inhibitors in Mesothelioma: For Want of Anything Better? J. Thorac. Oncol. 2017, 12, 778–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrò, L.; Morra, A.; Fonsatti, E.; Cutaia, O.; Amato, G.; Giannarelli, D.; Di Giacomo, A.M.; Danielli, R.; Altomonte, M.; Mutti, L.; et al. Tremelimumab for patients with chemotherapy-resistant advanced malignant mesothelioma: An open-label, single-arm, phase 2 trial. Lancet Oncol. 2013, 14, 1104–1111. [Google Scholar] [CrossRef]
- Maio, M.; Scherpereel, A.; Calabrò, L.; Aerts, J.; Cedres Perez, S.; Bearz, A.; Nackaerts, K.; Fennell, D.A.; Kowalski, D.; Tsao, A.S.; et al. Tremelimumab as second-line or third-line treatment in relapsed malignant mesothelioma (DETERMINE): A multicentre, international, randomised, double-blind, placebo-controlled phase 2b trial. Lancet Oncol. 2017, 18, 1261–1273. [Google Scholar] [CrossRef]
- Alley, E.W.; Lopez, J.; Santoro, A.; Morosky, A.; Saraf, S.; Piperdi, B.; van Brummelen, E. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): Preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2017, 18, 623–630. [Google Scholar] [CrossRef]
- Desai, A.; Karrison, T.; Rose, B.; Tan, Y.; Hill, B.; Pemberton, E.; Straus, C.; Seiwert, T.; Kindler, H.L. OA08.03 Phase II Trial of Pembrolizumab (NCT02399371) In Previously-Treated Malignant Mesothelioma (MM): Final Analysis. J. Thorac. Oncol. 2018, 13, S339. [Google Scholar] [CrossRef] [Green Version]
- Popat, S.; Curioni-Fontecedro, A.; Polydoropoulou, V.; Shah, R.; O’Brien, M.; Pope, A.; Fisher, P.; Spicer, J.; Roy, A.; Gilligan, D.; et al. LBA91_PRA multicentre randomized phase III trial comparing pembrolizumab (P) vs single agent chemotherapy (CT) for advanced pre-treated malignant pleural mesothelioma (MPM): Results from the European Thoracic Oncology Platform (ETOP 9-15) PROMISE-meso trial. Ann. Oncol. 2019, 30, v931. [Google Scholar]
- Fennell, D.A.; Kirkpatrick, E.; Cozens, K.; Nye, M.; Lester, J.; Hanna, G.; Steele, N.; Szlosarek, P.; Danson, S.; Lord, J.; et al. CONFIRM: A double-blind, placebo-controlled phase III clinical trial investigating the effect of nivolumab in patients with relapsed mesothelioma: Study protocol for a randomised controlled trial. Trials 2018, 19, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quispel-Janssen, J.; Zago, G.; Schouten, R.; Buikhuisen, W.; Monkhorst, K.; Thunissen, E.; Baas, P. OA13.01 A Phase II Study of Nivolumab in Malignant Pleural Mesothelioma (NivoMes): With Translational Research (TR) Biopies. J. Thorac. Oncol. 2017, 12, S292–S293. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Kijima, T.; Aoe, K.; Kato, T.; Fujimoto, N.; Nakagawa, K.; Takeda, Y.; Hida, T.; Kanai, K.; Imamura, F.; et al. Clinical Efficacy and Safety of Nivolumab: Results of a Multicenter, Open-label, Single-arm, Japanese Phase II study in Malignan Pleural Mesothelioma (MERIT). Clin. Cancer Res. 2019, 25, 5485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forde, P.M.; Scherpereel, A.; Tsao, A.S. Use of Immune Checkpoint Inhibitors in Mesothelioma. Curr. Treat. Options Oncol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.K.; Lesterhuis, W.J.; Hughes, B.G.M.; Brown, C.; Kok, P.S.; O’Byrne, K.J.; John, T.; Pavlakis, N.; Kao, S.C.-H.; Yip, S.; et al. DREAM: A phase II study of durvalumab with first line chemotherapy in mesothelioma—First results. JCO 2018, 36, 8503-8503. [Google Scholar] [CrossRef]
- Calabrò, L.; Morra, A.; Giannarelli, D.; Amato, G.; D’Incecco, A.; Covre, A.; Lewis, A.; Rebelatto, M.C.; Danielli, R.; Altomonte, M.; et al. Tremelimumab combined with durvalumab in patients with mesothelioma (NIBIT-MESO-1): An open-label, non-randomised, phase 2 study. Lancet Respir. Med. 2018, 6, 451–460. [Google Scholar] [CrossRef]
- Scherpereel, A.; Mazieres, J.; Greillier, L.; Lantuejoul, S.; Dô, P.; Bylicki, O.; Monnet, I.; Corre, R.; Audigier-Valette, C.; Locatelli-Sanchez, M.; et al. Nivolumab or nivolumab plus ipilimumab in patients with relapsed malignant pleural mesothelioma (IFCT-1501 MAPS2): A multicentre, open-label, randomised, non-comparative, phase 2 trial. Lancet Oncol. 2019, 20, 239–253. [Google Scholar] [CrossRef]
- Disselhorst, M.J.; Quispel-Janssen, J.; Lalezari, F.; Monkhorst, K.; de Vries, J.F.; van der Noort, V.; Harms, E.; Burgers, S.; Baas, P. Ipilimumab and nivolumab in the treatment of recurrent malignant pleural mesothelioma (INITIATE): Results of a prospective, single-arm, phase 2 trial. Lancet Respir. Med. 2019, 7, 260–270. [Google Scholar] [CrossRef]
- Zeltsman, M.; Dozier, J.; McGee, E.; Ngai, D.; Adusumilli, P.S. CAR T-cell therapy for lung cancer and malignant pleural mesothelioma. Transl. Res. 2017, 187, 1–10. [Google Scholar] [CrossRef]
- Martinez, M.; Moon, E.K. CAR T Cells for Solid Tumors: New Strategies for Finding, Infiltrating, and Surviving in the Tumor Microenvironment. Front. Immunol. 2019, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Lv, J.; Li, P. Mesothelin as a biomarker for targeted therapy. Biomark Res. 2019, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Moon, E.; Carpenito, C.; Paulos, C.M.; Liu, X.; Brennan, A.L.; Chew, A.; Carroll, R.G.; Scholler, J.; Levine, B.L.; et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 2010, 70, 9053–9061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Jiang, D.; Yang, H.; He, Z.; Liu, X.; Qin, W.; Li, L.; Wang, C.; Li, Y.; Li, H.; et al. Modified CAR T cells targeting membrane-proximal epitope of mesothelin enhances the antitumor function against large solid tumor. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adusumilli, P.S.; Zauderer, M.G.; Rusch, V.W.; O’Cearbhaill, R.; Zhu, A.; Ngai, D.; McGee, E.; Chintala, N.; Messinger, J.; Cheema, W.; et al. Regional delivery of mesothelin-targeted CAR T cells for pleural cancers: Safety and preliminary efficacy in combination with anti-PD-1 agent. JCO 2019, 37, 2511-2511. [Google Scholar] [CrossRef]
- Adusumilli, P.S.; Zauderer, M.G.; Rusch, V.W.; O 039 Cearbhaill, R.E.; Zhu, A.; Ngai, D.A.; McGee, E.; Chintala, N.K.; Messinger, J.C.; Vincent, A.; et al. Abstract CT036: A phase I clinical trial of malignant pleural disease treated with regionally delivered autologous mesothelin-targeted CAR T cells: Safety and efficacy. Cancer Res. 2019, 79, CT036. [Google Scholar]
- De Gooijer, C.J.; Borm, F.J.; Scherpereel, A.; Baas, P. Immunotherapy in Malignant Pleural Mesothelioma. Front. Oncol. 2020, 10, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barsky, A.R.; Cengel, K.A.; Katz, S.I.; Sterman, D.H.; Simone, C.B. First-ever Abscopal Effect after Palliative Radiotherapy and Immuno-gene Therapy for Malignant Pleural Mesothelioma. Cureus 2019, 11, e4102. [Google Scholar] [CrossRef] [Green Version]
- Kepp, O.; Galluzzi, L.; Martins, I.; Schlemmer, F.; Adjemian, S.; Michaud, M.; Sukkurwala, A.Q.; Menger, L.; Zitvogel, L.; Kroemer, G. Molecular determinants of immunogenic cell death elicited by anticancer chemotherapy. Cancer Metastasis Rev. 2011, 30, 61–69. [Google Scholar] [CrossRef]
- Galluzzi, L.; Kepp, O.; Kroemer, G. Immunogenic cell death in radiation therapy. Oncoimmunology 2014, 2, e26536-3. [Google Scholar] [CrossRef] [Green Version]
- Zelenay, S.; Reis e Sousa, C. Adaptive immunity after cell death. Trends Immunol. 2013, 34, 329–335. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuodeh, Y.; Venkat, P.; Kim, S. Systematic review of case reports on the abscopal effect. Curr. Probl. Cancer 2016, 40, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wu, M.O.; De la Maza, L.; Yun, Z.; Yu, J.; Zhao, Y.; Cho, J.; de Perrot, M. Targeting the inhibitory receptor CTLA-4 on T cells increased abscopal effects in murine mesothelioma model. Oncotarget 2015, 6, 12468–12480. [Google Scholar] [PubMed] [Green Version]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Drayton, D.L.; Liao, S.; Mounzer, R.H.; Ruddle, N.H. Lymphoid organ development: From ontogeny to neogenesis. Nat. Immunol. 2006, 7, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Dieu-Nosjean, M.-C.; Giraldo, N.A.; Kaplon, H.; Germain, C.; Fridman, W.H.; Sautès-Fridman, C. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol. Rev. 2016, 271, 260–275. [Google Scholar] [CrossRef]
- Colbeck, E.J.; Ager, A.; Gallimore, A.; Jones, G.W. Tertiary Lymphoid Structures in Cancer: Drivers of Antitumor Immunity, Immunosuppression, or Bystander Sentinels in Disease? Front. Immunol. 2017, 8, 1830. [Google Scholar] [CrossRef] [Green Version]
- Sautès-Fridman, C.; Lawand, M.; Giraldo, N.A.; Kaplon, H.; Germain, C.; Fridman, W.H.; Dieu-Nosjean, M.-C. Tertiary Lymphoid Structures in Cancers: Prognostic Value, Regulation, and Manipulation for Therapeutic Intervention. Front. Immunol. 2016, 7, 407. [Google Scholar] [CrossRef] [Green Version]
- Martinet, L.; Garrido, I.; Filleron, T.; Le Guellec, S.; Bellard, E.; Fournie, J.J.; Rochaix, P.; Girard, J.P. Human Solid Tumors Contain High Endothelial Venules: Association with T- and B-Lymphocyte Infiltration and Favorable Prognosis in Breast Cancer. Cancer Res. 2011, 71, 5678–5687. [Google Scholar] [CrossRef] [Green Version]
- Germain, C.; Gnjatic, S.; Tamzalit, F.; Knockaert, S.; Remark, R.; Goc, J.; Lepelley, A.; Becht, E.; Katsahian, S.; Bizouard, G.; et al. Presence of B Cells in Tertiary Lymphoid Structures Is Associated with a Protective Immunity in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 189, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, D.S.; Wood, D.E.; Akerley, W.; Bazhenova, L.A.; Borghaei, H.; Camidge, D.R.; Cheney, R.T.; Chirieac, L.R.; D’Amico, T.A.; Dilling, T.; et al. NCCN Guidelines Insights: Malignant Pleural Mesothelioma, Version 3.2016. J. Natl. Compr. Cancer Netw. 2016, 14, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.; Oizumi, S.; Kikuchi, E.; Shinagawa, N.; Konishi-Sakakibara, J.; Ishimine, A.; Aoe, K.; Gemba, K.; Kishimoto, T.; Torigoe, T.; et al. CD8+ tumor-infiltrating lymphocytes predict favorable prognosis in malignant pleural mesothelioma after resection. Cancer Immunol. Immunother. 2010, 59, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Anraku, M.; Cunningham, K.S.; Yun, Z.; Tsao, M.-S.; Zhang, L.; Keshavjee, S.; Johnston, M.R.; de Perrot, M. Impact of tumor-infiltrating T cells on survival in patients with malignant pleural mesothelioma. J. Thorac. Cardiovasc. Surg. 2008, 135, 823–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, K.; Kadota, K.; Sima, C.S.; Sadelain, M.; Rusch, V.W.; Travis, W.D.; Adusumilli, P.S. Chronic inflammation in tumor stroma is an independent predictor of prolonged survival in epithelioid malignant pleural mesothelioma patients. Cancer Immunol. Immunother. 2011, 60, 1721–1728. [Google Scholar] [CrossRef]
- Goc, J.; Germain, C.; Vo-Bourgais, T.K.D.; Lupo, A.; Klein, C.; Knockaert, S.; de Chaisemartin, L.; Ouakrim, H.; Becht, E.; Alifano, M.; et al. Dendritic Cells in Tumor-Associated Tertiary Lymphoid Structures Signal a Th1 Cytotoxic Immune Contexture and License the Positive Prognostic Value of Infiltrating CD8+ T Cells. Cancer Res. 2014, 74, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieu-Nosjean, M.-C.; Antoine, M.; Danel, C.; Heudes, D.; Wislez, M.; Poulot, V.; Rabbe, N.; Laurans, L.; Tartour, E.; de Chaisemartin, L.; et al. Long-Term Survival for Patients With Non–Small-Cell Lung Cancer With Intratumoral Lymphoid Structures. JCO 2008, 26, 4410–4417. [Google Scholar] [CrossRef] [Green Version]
- Silina, K.; Soltermann, A.; Attar, F.M.; Casanova, R.; Uckeley, Z.M.; Thut, H.; Wandres, M.; Isajevs, S.; Cheng, P.; Curioni-Fontecedro, A.; et al. Germinal Centers Determine the Prognostic Relevance of Tertiary Lymphoid Structures and Are Impaired by Corticosteroids in Lung Squamous Cell Carcinoma. Cancer Res. 2018, 78, 1308–1320. [Google Scholar] [CrossRef] [Green Version]
- Di Caro, G.; Bergomas, F.; Grizzi, F.; Doni, A.; Bianchi, P.; Malesci, A.; Laghi, L.; Allavena, P.; Mantovani, A.; Marchesi, F. Occurrence of Tertiary Lymphoid Tissue Is Associated with T-Cell Infiltration and Predicts Better Prognosis in Early-Stage Colorectal Cancers. Clin. Cancer Res. 2014, 20, 2147–2158. [Google Scholar] [CrossRef] [Green Version]
- Posch, F.; Silina, K.; Leibl, S.; Mündlein, A.; Moch, H.; Siebenhüner, A.; Samaras, P.; Riedl, J.; Stotz, M.; Szkandera, J.; et al. Maturation of tertiary lymphoid structures and recurrence of stage II and III colorectal cancer. Oncoimmunology 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hiraoka, N.; Ino, Y.; Yamazaki-Itoh, R.; Kanai, Y.; Kosuge, T.; Shimada, K. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br. J. Cancer 2015, 112, 1782–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castino, G.F.; Cortese, N.; Capretti, G.; Serio, S.; Di Caro, G.; Mineri, R.; Magrini, E.; Grizzi, F.; Cappello, P.; Novelli, F.; et al. Spatial distribution of B cells predicts prognosis in human pancreatic adenocarcinoma. Oncoimmunology 2016, 5, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirsing, A.M.; Ervik, I.K.; Seppola, M.; Uhlin-Hansen, L.; Steigen, S.E.; Hadler-Olsen, E. Presence of high-endothelial venules correlates with a favorable immune microenvironment in oral squamous cell carcinoma. Mod. Pathol. 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; de Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, J.Y.; Park, I.A.; Song, I.H.; Yu, J.H.; Ahn, J.-H.; Gong, G. Prognostic Significance of Tumor-Infiltrating Lymphocytes and the Tertiary Lymphoid Structures in HER2-Positive Breast Cancer Treated With Adjuvant Trastuzumab. Am. J. Clin. Pathol. 2015, 144, 278–288. [Google Scholar] [CrossRef] [Green Version]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef]
- Colbeck, E.J.; Jones, E.; Hindley, J.P.; Smart, K.; Schulz, R.; Browne, M.; Cutting, S.; Williams, A.; Parry, L.; Godkin, A.; et al. Treg Depletion Licenses T Cell-Driven HEV Neogenesis and Promotes Tumor Destruction. Cancer Immunol. Res. 2017, 5, 1005–1015. [Google Scholar] [CrossRef] [Green Version]
- Joshi, N.S.; Akama-Garren, E.H.; Lu, Y.; Lee, D.-Y.; Chang, G.P.; Li, A.; DuPage, M.; Tammela, T.; Kerper, N.R.; Farago, A.F.; et al. Regulatory T Cells in Tumor-Associated Tertiary Lymphoid Structures Suppress Anti-tumor T Cell Responses. Immunity 2015, 43, 579–590. [Google Scholar] [CrossRef] [Green Version]
- Fear, V.S.; Tilsed, C.; Chee, J.; Forbes, C.A.; Casey, T.; Solin, J.N.; Lansley, S.M.; Lesterhuis, W.J.; Dick, I.M.; Nowak, A.K.; et al. Combination immune checkpoint blockade as an effective therapy for mesothelioma. Oncoimmunology 2018, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Allen, E.; Jabouille, A.; Rivera, L.B.; Lodewijckx, I.; Missiaen, R.; Steri, V.; Feyen, K.; Tawney, J.; Hanahan, D.; Michael, I.P.; et al. Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci. Transl. Med. 2017, 9, eaak9679. [Google Scholar] [CrossRef] [Green Version]
- Coppola, D.; Nebozhyn, M.; Khalil, F.; Dai, H.; Yeatman, T.; Loboda, A.; Mulé, J.J. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am. J. Pathol. 2011, 179, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Sautès-Fridman, C.; Petitprez, F.; Calderaro, J.; Fridman, W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Publ. Group 2019, 19, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Montfort, A.; Pearce, O.; Maniati, E.; Vincent, B.G.; Bixby, L.; Böhm, S.; Dowe, T.; Wilkes, E.H.; Chakravarty, P.; Thompson, R.; et al. A Strong B-cell Response Is Part of the Immune Landscape in Human High-Grade Serous Ovarian Metastases. Clin. Cancer Res. 2017, 23, 250–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Hernández, M.D.L.L.; Uribe-Uribe, N.O.; Espinosa-González, R.; Kast, W.M.; Khader, S.A.; Rangel-Moreno, J. A Unique Cellular and Molecular Microenvironment Is Present in Tertiary Lymphoid Organs of Patients with Spontaneous Prostate Cancer Regression. Front. Immunol. 2017, 8, 563. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, A.S.; Peikert, T.; Smadbeck, J.B.; Udell, J.B.M.; Garcia-Rivera, E.; Elsbernd, L.; Erskine, C.L.; Van Keulen, V.P.; Kosari, F.; Murphy, S.J.; et al. Neoantigenic Potential of Complex Chromosomal Rearrangements in Mesothelioma. J. Thorac. Oncol. 2019, 14, 276–287. [Google Scholar] [CrossRef] [Green Version]
- Kiyotani, K.; Park, J.-H.; Inoue, H.; Husain, A.; Olugbile, S.; Zewde, M.; Nakamura, Y.; Vigneswaran, W.T. Integrated analysis of somatic mutations and immune microenvironment in malignant pleural mesothelioma. Oncoimmunology 2017, 6, e1278330. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.S.; Sahota, R.A.; Milne, K.; Kost, S.E.; Nesslinger, N.J.; Watson, P.H.; Nelson, B.H. CD20+ Tumor-Infiltrating Lymphocytes Have an Atypical CD27- Memory Phenotype and Together with CD8+ T Cells Promote Favorable Prognosis in Ovarian Cancer. Clin. Cancer Res. 2012, 18, 3281–3292. [Google Scholar] [CrossRef] [Green Version]
- Kroeger, D.R.; Milne, K.; Nelson, B.H. Tumor-Infiltrating Plasma Cells Are Associated with Tertiary Lymphoid Structures, Cytolytic T-Cell Responses, and Superior Prognosis in Ovarian Cancer. Clin. Cancer Res. 2016, 22, 3005–3015. [Google Scholar] [CrossRef] [Green Version]
- Coronella, J.A.; Spier, C.; Welch, M.; Trevor, K.T.; Stopeck, A.T.; Villar, H.; Hersh, E.M. Antigen-Driven Oligoclonal Expansion of Tumor-Infiltrating B Cells in Infiltrating Ductal Carcinoma of the Breast. J. Immunol. 2002, 169, 1829–1836. [Google Scholar] [CrossRef] [Green Version]
- Schlößer, H.A.; Thelen, M.; Lechner, A.; Wennhold, K.; Garcia-Marquez, M.A.; Rothschild, S.I.; Staib, E.; Zander, T.; Beutner, D.; Gathof, B.; et al. B cells in esophago-gastric adenocarcinoma are highly differentiated, organize in tertiary lymphoid structures and produce tumor-specific antibodies. Oncoimmunology 2018, 8, e1512458. [Google Scholar] [CrossRef] [Green Version]
- Nzula, S.; Going, J.J.; Stott, D.I. Antigen-driven clonal proliferation, somatic hypermutation, and selection of B lymphocytes infiltrating human ductal breast carcinomas. Cancer Res. 2003, 63, 3275–3280. [Google Scholar] [PubMed]
- Cipponi, A.; Mercier, M.; Seremet, T.; Baurain, J.-F.; Théate, I.; van den Oord, J.; Stas, M.; Boon, T.; Coulie, P.G.; van Baren, N. Neogenesis of Lymphoid Structures and Antibody Responses Occur in Human Melanoma Metastases. Cancer Res. 2012, 72, 3997–4007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wouters, M.C.A.; Nelson, B.H. Prognostic Significance of Tumor-Infiltrating B Cells and Plasma Cells in Human Cancer. Clin. Cancer Res. 2018, 24, 6125–6135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petitprez, F.; de Reyniès, A.; Keung, E.Z.; Chen, T.W.-W.; Sun, C.-M.; Calderaro, J.; Jeng, Y.-M.; Hsiao, L.-P.; Lacroix, L.; Bougoüin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Skaarup Larsen, M.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Krishnan, S.; Bakker, E.; Lee, C.; Kissick, H.T.; Ireland, D.J.; Beilharz, M.W. Successful combined intratumoral immunotherapy of established murine mesotheliomas requires B-cell involvement. J. Interferon Cytokine Res. 2015, 35, 100–107. [Google Scholar] [CrossRef]
- Ujiie, H.; Kadota, K.; Nitadori, J.-I.; Aerts, J.G.; Woo, K.M.; Sima, C.S.; Travis, W.D.; Jones, D.R.; Krug, L.M.; Adusumilli, P.S. The tumoral and stromal immune microenvironment in malignant pleural mesothelioma: A comprehensive analysis reveals prognostic immune markers. Oncoimmunology 2015, 4, e1009285. [Google Scholar] [CrossRef] [Green Version]
- Minnema-Luiting, J.; Vroman, H.; Aerts, J.; Cornelissen, R. Heterogeneity in Immune Cell Content in Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2018, 19, 1041. [Google Scholar] [CrossRef] [Green Version]
- Chee, S.J.; Lopez, M.; Mellows, T.; Gankande, S.; Moutasim, K.A.; Harris, S.; Clarke, J.; Vijayanand, P.; Thomas, G.J.; Ottensmeier, C.H. Evaluating the effect of immune cells on the outcome of patients with mesothelioma. Br. J. Cancer 2017, 117, 1341–1348. [Google Scholar] [CrossRef]
- Jackaman, C.; Cornwall, S.; Graham, P.T.; Nelson, D.J. CD40-activated B cells contribute to mesothelioma tumor regression. Immunol. Cell Biol. 2011, 89, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Kahles, A.; Lehmann, K.-V.; Toussaint, N.C.; Hüser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Cancer Genome Atlas Research Network; et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell 2018, 34, 211–224.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGranahan, N.; Furness, A.J.S.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dantas-Barbosa, C.; de Macedo Brigido, M.; Maranhao, A.Q. Antibody phage display libraries: Contributions to oncology. Int. J. Mol. Sci. 2012, 13, 5420–5440. [Google Scholar] [CrossRef]
- Frenzel, A.; Schirrmann, T.; Hust, M. Phage display-derived human antibodies in clinical development and therapy. MAbs 2016, 8, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Rouet, R.; Jackson, K.J.L.; Langley, D.B.; Christ, D. Next-Generation Sequencing of Antibody Display Repertoires. Front. Immunol. 2018, 9, 1315-1315. [Google Scholar] [CrossRef] [Green Version]
- An, F.; Drummond, D.C.; Wilson, S.; Kirpotin, D.B.; Nishimura, S.L.; Broaddus, V.C.; Liu, B. Targeted drug delivery to mesothelioma cells using functionally selected internalizing human single-chain antibodies. Mol. Cancer Ther. 2008, 7, 569–578. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Guan, C.-W.; Song, Y.; Wang, H. The multifaceted role of CD146/MCAM in the promotion of melanoma progression. Cancer Cell Int. 2015, 15, 3–11. [Google Scholar] [CrossRef] [Green Version]
- De Kruijff, I.; Timmermans, A.; den Bakker, M.; Trapman-Jansen, A.; Foekens, R.; Meijer-Van Gelder, M.; Oomen-de Hoop, E.; Smid, M.; Hollestelle, A.; van Deurzen, C.; et al. The Prevalence of CD146 Expression in Breast Cancer Subtypes and Its Relation to Outcome. Cancers 2018, 10, 134. [Google Scholar] [CrossRef] [Green Version]
- Olajuyin, A.M.; Olajuyin, A.K.; Wang, Z.; Zhao, X.; Zhang, X. CD146 T cells in lung cancer: Its function, detection, and clinical implications as a biomarker and therapeutic target. Cancer Cell Int. 2019, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bidlingmaier, S.; He, J.; Wang, Y.; An, F.; Feng, J.; Barbone, D.; Gao, D.; Franc, B.; Broaddus, V.C.; Liu, B. Identification of MCAM/CD146 as the target antigen of a human monoclonal antibody that recognizes both epithelioid and sarcomatoid types of mesothelioma. Cancer Res. 2009, 69, 1570–1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beije, N.; Kraan, J.; Bakker, M.A.; Maat, A.P.W.M.; Leest, C.; Cornelissen, R.; Van, N.M.; Martens, J.W.M.; Aerts, J.G.J.V.; Sleijfer, S. Improved diagnosis and prognostication of patients with pleural malignant mesothelioma using biomarkers in pleural effusions and peripheral blood samples—A short report. Cell. Oncol. 2017, 40, 511–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbas, C.F.; Kang, A.S.; Lerner, R.A.; Benkovic, S.J. Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc. Natl. Acad. Sci. USA 1991, 88, 7978–7982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Garen, A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: Selection of specific antibodies from single-chain Fv fusion phage libraries. Proc. Natl. Acad. Sci. USA 1995, 92, 6537–6541. [Google Scholar] [CrossRef] [Green Version]
- Wortzel, R.D.; Urban, J.L.; Philipps, C.; Fitch, F.W.; Schreiber, H. Independent immunodominant and immunorecessive tumor-specific antigens on a malignant tumor: Antigenic dissection with cytolytic T cell clones. J. Immunol. 1983, 130, 2461–2466. [Google Scholar]
- Brüggemann, M.; Osborn, M.J.; Ma, B.; Hayre, J.; Avis, S.; Lundstrom, B.; Buelow, R. Human antibody production in transgenic animals. Arch. Immunol. Exp. 2015, 63, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.-C.; Liang, Q.; Ali, H.; Bayliss, L.; Beasley, A.; Bloomfield-Gerdes, T.; Bonoli, L.; Brown, R.; Campbell, J.; Carpenter, A.; et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014, 32, 356–363. [Google Scholar] [CrossRef]
- Jakobovits, A.; Amado, R.G.; Yang, X.; Roskos, L.; Schwab, G. From XenoMouse technology to panitumumab, the first fully human antibody product from transgenic mice. Nat. Biotechnol. 2007, 25, 1134–1143. [Google Scholar] [CrossRef]
- Lonberg, N. Fully human antibodies from transgenic mouse and phage display platforms. Curr. Opin. Immunol. 2008, 20, 450–459. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Mose, L.E.; Selitsky, S.R.; Bixby, L.M.; Marron, D.L.; Iglesia, M.D.; Serody, J.S.; Perou, C.M.; Vincent, B.G.; Parker, J.S. Assembly-based inference of B-cell receptor repertoires from short read RNA sequencing data with V’DJer. Bioinformatics 2016, 32, 3729–3734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selitsky, S.R.; Mose, L.E.; Smith, C.C.; Chai, S.; Hoadley, K.A.; Dittmer, D.P.; Moschos, S.J.; Parker, J.S.; Vincent, B.G. Prognostic value of B cells in cutaneous melanoma. Genome Med. 2019, 11, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Traggiai, E.; Becker, S.; Subbarao, K.; Kolesnikova, L.; Uematsu, Y.; Gismondo, M.R.; Murphy, B.R.; Rappuoli, R.; Lanzavecchia, A. An efficient method to make human monoclonal antibodies from memory B cells: Potent neutralization of SARS coronavirus. Nat. Med. 2004, 10, 871–875. [Google Scholar] [CrossRef]
- Kwakkenbos, M.J.; van Helden, P.M.; Beaumont, T.; Spits, H. Stable long-term cultures of self-renewing B cells and their applications. Immunol. Rev. 2016, 270, 65–77. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, A.E.; Karagiannis, P.; Dodev, T.; Koers, A.; Lacy, K.; Josephs, D.H.; Takhar, P.; Geh, J.L.C.; Healy, C.; Harries, M.; et al. Monitoring the Systemic Human Memory B Cell Compartment of Melanoma Patients for Anti-Tumor IgG Antibodies. PLoS ONE 2011, 6, e19330. [Google Scholar] [CrossRef]
- Clargo, A.M.; Hudson, A.R.; Ndlovu, W.; Wootton, R.J.; Cremin, L.A.; O’Dowd, V.L.; Nowosad, C.R.; Starkie, D.O.; Shaw, S.P.; Compson, J.E.; et al. The rapid generation of recombinant functional monoclonal antibodies from individual, antigen-specific bone marrow-derived plasma cells isolated using a novel fluorescence-based method. MAbs 2014, 6, 143–159. [Google Scholar] [CrossRef] [Green Version]
- Tickle, S.; Howells, L.; O’Dowd, V.; Starkie, D.; Whale, K.; Saunders, M.; Lee, D.; Lightwood, D. A fully automated primary screening system for the discovery of therapeutic antibodies directly from B cells. J. Biomol. Screen 2015, 20, 492–497. [Google Scholar] [CrossRef] [Green Version]
- Jin, A.; Ozawa, T.; Tajiri, K.; Obata, T.; Kondo, S.; Kinoshita, K.; Kadowaki, S.; Takahashi, K.; Sugiyama, T.; Kishi, H.; et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat. Med. 2009, 15, 1088–1092. [Google Scholar] [CrossRef]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- Walker, L.M.; Phogat, S.K.; Chan-Hui, P.-Y.; Wagner, D.; Phung, P.; Goss, J.L.; Wrin, T.; Simek, M.D.; Fling, S.; Mitcham, J.L.; et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009, 326, 285–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turchaninova, M.A.; Davydov, A.; Britanova, O.V.; Shugay, M.; Bikos, V.; Egorov, E.S.; Kirgizova, V.I.; Merzlyak, E.M.; Staroverov, D.B.; Bolotin, D.A.; et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat. Protoc. 2016, 11, 1599–1616. [Google Scholar] [CrossRef] [PubMed]


| Screening Strategy | Approach | Antigen Display | Advantages | Disadvantages |
|---|---|---|---|---|
| Antigen-based screening (pre-selection of specificity based on antigen expression) | Phage-display technology with human antibody synthetic libraries [115,116] | In vitro adsorbed antigen |
|
|
| Expression of the antigen on the host cell membrane (biopanning) | ||||
| Transgenic mice expressing fully human antibodies [126,127,128] | Vaccination with soluble antigen |
|
| |
| MBC immortalization via BCL6/BCL-XL expression [134,135] | Soluble and fluorescently labeled antigen |
|
| |
| Antigen-unbiased screening (selection based on binding to cancer cells) | Phage-display technology with human antibody synthetic libraries [115,116,117] | Antigen on cell surface |
|
|
| BCR repertoire from the peripheral blood of elite responders pre- and post-therapy [142] | Antigen on cell surface |
|
| |
| Bioinformatic analysis of BCR repertoire in tumor tissue [130,131] | Antigen on cell surface |
|
| |
| EBV infection [134] | Antigen on cell surface |
|
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolini, F.; Bocchini, M.; Angeli, D.; Bronte, G.; Delmonte, A.; Crinò, L.; Mazza, M. Fully Human Antibodies for Malignant Pleural Mesothelioma Targeting. Cancers 2020, 12, 915. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12040915
Nicolini F, Bocchini M, Angeli D, Bronte G, Delmonte A, Crinò L, Mazza M. Fully Human Antibodies for Malignant Pleural Mesothelioma Targeting. Cancers. 2020; 12(4):915. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12040915
Chicago/Turabian StyleNicolini, Fabio, Martine Bocchini, Davide Angeli, Giuseppe Bronte, Angelo Delmonte, Lucio Crinò, and Massimiliano Mazza. 2020. "Fully Human Antibodies for Malignant Pleural Mesothelioma Targeting" Cancers 12, no. 4: 915. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12040915