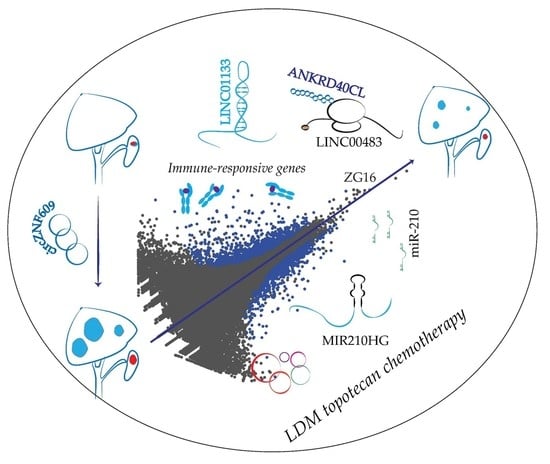
Gene Expression Signatures of a Preclinical Mouse Model during Colorectal Cancer Progression under Low-Dose Metronomic Chemotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. A mouse Model for Profiling Dysregulated Genes during LDM Topotecan Chemotherapy
2.2. MiRNA-Seq Revealed miRNA Dysregulation during LDM Topotecan Chemotherapy
2.3. Protein Coding Gene Expression Analyses Identify Several Immune-Regulated Genes Affected by LDM Topotecan Treatment
2.4. LncRNA Expression Analysis Identifies LINC01133 and LINC00483 as Potential LDM Topotecan Chemotherapy Targets
2.5. circZNF609 as Upregulated circRNA during Cancer Progression
2.6. CircZNF609 Promotes Colorectal Cancer Progression in Mouse Xenografts
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Antibodies
4.3. Cell Lines, Cell Culture and Generation of Stable Cell Lines
4.4. Mouse Strains
4.5. Intrasplenic Implantation
4.6. Subcutaneous Implantation
4.7. Animal Handling
4.8. Preparation of Small RNA Library and Data Analysis
4.9. Preparation of rRNA-Depleted Library and Analysis of mRNAs and lncRNAs
4.10. circRNA Detection and Differential Expression Analysis
4.11. Validation of circRNAs Expression by qPCR
4.12. Proliferation Assay
4.13. Nuclear and Cytoplasmic Fractionation
4.14. SDS-PAGE and Western Blotting
4.15. Preparation of RNA
4.16. Northern Blotting
4.17. Ago2 Immunoprecipitations (IP)
4.18. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kerbel, R.S.; Kamen, B.A. The anti-angiogenic basis of metronomic chemotherapy. Nat. Rev. Cancer 2004, 4, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, E.; Kavallaris, M.; André, N. Metronomic chemotherapy: New rationale for new directions. Nat. Rev. Clin. Oncol. 2010, 7, 455–465. [Google Scholar] [CrossRef]
- Mutsaers, A.J. Metronomic Chemotherapy. Top. Companion Anim. Med. 2009, 24, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Kerbel, R.S.; Shaked, Y. The potential clinical promise of ‘multimodality’ metronomic chemotherapy revealed by preclinical studies of metastatic disease. Cancer Lett. 2017, 400, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Muñoz, W.; Di Desidero, T.; Man, S.; Xu, P.; Jaramillo, M.L.; Hashimoto, K.; Collins, C.; Banville, M.; O’Connor-Mccourt, M.D.; Kerbel, R.S. Analysis of acquired resistance to metronomic oral topotecan chemotherapy plus pazopanib after prolonged preclinical potent responsiveness in advanced ovarian cancer. Angiogenesis 2014, 17, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, M.E.; Biganzoli, L.; Cortesi, L.; De Placido, S.; Donadio, M.; Fabi, A.; Ferro, A.; Generali, D.; Lorusso, V.; Milani, A.; et al. Treating advanced breast cancer with metronomic chemotherapy: What is known, what is new and what is the future? Onco. Targets. Ther. 2019, 12, 2989–2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simsek, C.; Esin, E.; Yalcin, S. Metronomic Chemotherapy: A Systematic Review of the Literature and Clinical Experience. J. Oncol. 2019, 2019. [Google Scholar] [CrossRef] [Green Version]
- Riesco-Martinez, M.; Parra, K.; Saluja, R.; Francia, G.; Emmenegger, U. Resistance to metronomic chemotherapy and ways to overcome it. Cancer Lett. 2017, 400, 311–318. [Google Scholar] [CrossRef] [Green Version]
- Klement, G.; Baruchel, S.; Rak, J.; Man, S.; Clark, K.; Hicklin, D.J.; Bohlen, P.; Kerbel, R.S. Continuous low-dose therapy with vinblastine and VEGF receptor-2 antibody induces sustained tumor regression without overt toxicity. J. Clin. Investig. 2000, 105, R15–R24. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.A.; Ho, C.M.; Chang, M.C.; Sun, W.Z.; Chen, Y.L.; Chiang, Y.C.; Syu, M.H.; Hsieh, C.Y.; Cheng, W.F. Metronomic chemotherapy enhances antitumor effects of cancer vaccine by depleting regulatory T lymphocytes and inhibiting tumor angiogenesis. Mol. Ther. 2010, 18, 1233–1243. [Google Scholar] [CrossRef]
- André, N.; Tsai, K.; Carré, M.; Pasquier, E. Metronomic Chemotherapy: Direct Targeting of Cancer Cells after all? Trends Cancer 2017, 3, 319–325. [Google Scholar] [CrossRef]
- Francia, G.; Shaked, Y.; Hashimoto, K.; Sun, J.; Yin, M.; Cesta, C.; Xu, P.; Man, S.; Hackl, C.; Stewart, J.; et al. Low-dose metronomic oral dosing of a prodrug of gemcitabine (LY2334737) causes antitumor effects in the absence of inhibition of systemic vasculogenesis. Mol. Cancer Ther. 2012, 11, 680–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gnoni, A.; Silvestris, N.; Licchetta, A.; Santini, D.; Scartozzi, M.; Ria, R.; Pisconti, S.; Petrelli, F.; Vacca, A.; Lorusso, V. Metronomic chemotherapy from rationale to clinical studies: A dream or reality? Crit. Rev. Oncol. Hematol. 2015, 95, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Chang, M.C.; Cheng, W.F. Metronomic chemotherapy and immunotherapy in cancer treatment. Cancer Lett. 2017, 400, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Simkens, L.H.J.; Van Tinteren, H.; May, A.; Ten Tije, A.J.; Creemers, G.J.M.; Loosveld, O.J.L.; De Jongh, F.E.; Erdkamp, F.L.G.; Erjavec, Z.; Van Der Torren, A.M.E.; et al. Maintenance treatment with capecitabine and bevacizumab in metastatic colorectal cancer (CAIRO3): A phase 3 randomised controlled trial of the Dutch Colorectal Cancer Group. Lancet 2015, 385, 1843–1852. [Google Scholar] [CrossRef]
- Tol, J.; Koopman, M.; Cats, A.; Rodenburg, C.J.; Creemers, G.J.M.; Schrama, J.G.; Erdkamp, F.L.G.; Vos, A.H.; van Groeningen, C.J.; Sinnige, H.A.M.; et al. Chemotherapy, Bevacizumab, and Cetuximab in Metastatic Colorectal Cancer. N. Engl. J. Med. 2009, 360, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Gustavsson, B.; Carlsson, G.; MacHover, D.; Petrelli, N.; Roth, A.; Schmoll, H.J.; Tveit, K.M.; Gibson, F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin. Colorectal Cancer 2015, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Woo, I.S.; Jung, Y.H. Metronomic chemotherapy in metastatic colorectal cancer. Cancer Lett. 2017, 400, 319–324. [Google Scholar] [CrossRef]
- Hackl, C.; Man, S.; Francia, G.; Milsom, C.; Xu, P.; Kerbel, R.S. Metronomic oral topotecan prolongs survival and reduces liver metastasis in improved preclinical orthotopic and adjuvant therapy colon cancer models. Gut 2013, 62, 259–271. [Google Scholar] [CrossRef]
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brazert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef] [Green Version]
- Świerczewska, M.; Klejewski, A.; Wojtowicz, K.; Brązert, M.; Iżycki, D.; Nowicki, M.; Zabel, M.; Januchowski, R. New and Old Genes Associated with Primary and Established Responses to Cisplatin and Topotecan Treatment in Ovarian Cancer Cell Lines. Molecules 2017, 22, 1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Fabbri, M.; Girnita, L.; Varani, G.; Calin, G.A. Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res. 2019, 29, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Pasut, A.; Matsumoto, A.; Clohessy, J.G.; Pandolfi, P.P. The pleiotropic role of non-coding genes in development and cancer. Curr. Opin. Cell Biol. 2016, 43, 104–113. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bester, A.C.; Lee, J.D.; Chavez, A.; Lee, Y.R.; Nachmani, D.; Vora, S.; Victor, J.; Sauvageau, M.; Monteleone, E.; Rinn, J.L.; et al. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell 2018, 173, 649–664.e20. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Liu, Q.; Li, C.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.L.; Wang, J.; Hu, H.; et al. LncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017, 23, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014, 281, 1750–1758. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. P53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Makarewich, C.A.; Olson, E.N. Mining for Micropeptides. Trends Cell Biol. 2017, 27, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Polycarpou-Schwarz, M.; Groß, M.; Mestdagh, P.; Schott, J.; Grund, S.E.; Hildenbrand, C.; Rom, J.; Aulmann, S.; Sinn, H.P.; Vandesompele, J.; et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 2018, 37, 4750–4768. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP 2A- BP encoded by LINC 00665 inhibits triple-negative breast cancer progression. EMBO J. 2020, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef] [Green Version]
- Li, X.L.; Pongor, L.; Tang, W.; Das, S.; Muys, B.R.; Jones, M.F.; Lazar, S.B.; Dangelmaier, E.A.; Hartford, C.C.; Grammatikakis, I.; et al. A small protein encoded by a putative lncrna regulates apoptosis and tumorigenicity in human colorectal cancer cells. Elife 2020, 9, 1–44. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [Green Version]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18. [Google Scholar] [CrossRef]
- Maass, P.G.; Glažar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Kadener, S. circRNAs in Cancer. Curr. Opin. Genet. Dev. 2018, 48, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019, 176, 831–843.e22. [Google Scholar] [CrossRef] [Green Version]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Yang, Q.; Dai, Y.; Guo, W.; Du, H.; Song, L.; Peng, X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-ΚB signaling pathway. Mol. Cancer 2017, 16. [Google Scholar] [CrossRef] [Green Version]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; Hausen, A.Z.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef]
- Bae, D.H.; Jansson, P.J.; Huang, M.L.; Kovacevic, Z.; Kalinowski, D.; Lee, C.S.; Sahni, S.; Richardson, D.R. The role of NDRG1 in the pathology and potential treatment of human cancers. J. Clin. Pathol. 2013, 66, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Cantelmo, A.R.; Conradi, L.C.; Brajic, A.; Goveia, J.; Kalucka, J.; Pircher, A.; Chaturvedi, P.; Hol, J.; Thienpont, B.; Teuwen, L.A.; et al. Inhibition of the Glycolytic Activator PFKFB3 in Endothelium Induces Tumor Vessel Normalization, Impairs Metastasis, and Improves Chemotherapy. Cancer Cell 2016, 30, 968–985. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Zhou, N.; Zhang, D.; Zhang, K.; Zheng, W.; Bao, Y.; Yang, W. PFKFB3 inhibition attenuates oxaliplatin-induced autophagy and enhances its cytotoxicity in colon cancer cells. Int. J. Mol. Sci. 2019, 20, 5415. [Google Scholar] [CrossRef] [Green Version]
- Wangpu, X.; Yang, X.; Zhao, J.; Lu, J.; Guan, S.; Lu, J.; Kovacevic, Z.; Liu, W.; Mi, L.; Jin, R.; et al. The metastasis suppressor, NDRG1, inhibits “stemness” of colorectal cancer via down-regulation of nuclear β-catenin and CD44. Oncotarget 2015, 6, 33893–33911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Yu, C.; Yang, X.; Hong, H.; Lu, J.; Hu, W.; Hao, X.; Li, S.; Aikemu, B.; Yang, G.; et al. N-myc downstream-regulated gene 1 inhibits the proliferation of colorectal cancer through emulative antagonizing NEDD4-mediated ubiquitylation of p21. J. Exp. Clin. Cancer Res. 2019, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef] [PubMed]
- Han, H.G.; Moon, H.W.; Jeon, Y.J. ISG15 in cancer: Beyond ubiquitin-like protein. Cancer Lett. 2018, 438, 52–62. [Google Scholar] [CrossRef]
- Meng, H.; Li, W.; Boardman, L.A.; Wang, L. Loss of ZG16 is associated with molecular and clinicopathological phenotypes of colorectal cancer. BMC Cancer 2018, 18. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Du, P.; She, J.; Cao, L.; Li, Y.; Xia, H. Loss of ZG16 is regulated by miR-196a and contributes to stemness and progression of colorectal cancer. Oncotarget 2016, 7, 86695–86703. [Google Scholar] [CrossRef] [Green Version]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Hämmerle, M.; Eißmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Groß, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [Green Version]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial–mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, F.; Legnini, I.; Megiorni, F.; Colantoni, A.; Santini, T.; Morlando, M.; Di Timoteo, G.; Dattilo, D.; Dominici, C.; Bozzoni, I. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene 2019, 38, 3843–3854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho-Xuan, H.; GlaÅar, P.; Latini, C.; Heizler, K.; Haase, J.; Hett, R.; Anders, M.; Weichmann, F.; Bruckmann, A.; Van Den Berg, D.; et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 2020, 48, 10368–10382. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.Y.; Zhang, Y.Y.; Hemann, C.; Mahoney, C.E.; Zweier, J.L.; Loscalzo, J. MicroRNA-210 Controls Mitochondrial Metabolism during Hypoxia by Repressing the Iron-Sulfur Cluster Assembly Proteins ISCU1/2. Cell Metab. 2009, 10, 273–284. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.T.; Giaccia, A.J. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Mol. Cell 2009, 35, 856–867. [Google Scholar] [CrossRef] [Green Version]
- Grosso, S.; Doyen, J.; Parks, S.K.; Bertero, T.; Paye, A.; Cardinaud, B.; Gounon, P.; Lacas-Gervais, S.; Noël, A.; Pouysségur, J.; et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013, 4, e544. [Google Scholar] [CrossRef]
- Pranzini, E.; Leo, A.; Rapizzi, E.; Ramazzotti, M.; Magherini, F.; Giovannelli, L.; Caselli, A.; Cirri, P.; Taddei, M.L.; Paoli, P. miR-210-3p mediates metabolic adaptation and sustains DNA damage repair of resistant colon cancer cells to treatment with 5-fluorouracil. Mol. Carcinog. 2019, 58, 2181–2192. [Google Scholar] [CrossRef]
- Voellenkle, C.; Garcia-Manteiga, J.M.; Pedrotti, S.; Perfetti, A.; De Toma, I.; Da Silva, D.; Maimone, B.; Greco, S.; Fasanaro, P.; Creo, P.; et al. Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci. Rep. 2016, 6, 24141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Dang, J.; Song, A.; Cui, X.; Ma, Z.; Zhang, Z. Identification of LINC01234 and MIR210HG as novel prognostic signature for colorectal adenocarcinoma. J. Cell. Physiol. 2019, 234, 6769–6777. [Google Scholar] [CrossRef]
- Kang, X.; Kong, F.; Huang, K.; Li, L.; Li, Z.; Wang, X.; Zhang, W.; Wu, X. LncRNA MIR210HG promotes proliferation and invasion of non-small cell lung cancer by upregulating methylation of CACNA2D2 promoter via binding to DNMT1. Onco. Targets. Ther. 2019, 12, 3779–3790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Galluzzi, L.; Kepp, O.; Smyth, M.J.; Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015, 15, 405–414. [Google Scholar] [CrossRef]
- Desai, S.D.; Wood, L.M.; Tsai, Y.-C.; Hsieh, T.-S.; Marks, J.R.; Scott, G.L.; Giovanella, B.C.; Liu, L.F. ISG15 as a novel tumor biomarker for drug sensitivity. Mol. Cancer Ther. 2008, 7, 1430–1439. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.J.Y.; Pung, Y.F.; Sze, N.S.K.; Chin, K.C. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. USA 2006, 103, 10735–10740. [Google Scholar] [CrossRef] [Green Version]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Piao, H.L.; Kim, B.J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; MacKowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Yang, M.; Liao, A.; Zeng, B.; Liu, D.; Yao, Y.; Hu, G.; Chen, X.; Feng, Z.; Du, Y.; et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J. Cell. Mol. Med. 2018, 22, 3875–3886. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, Z.; Qin, B. A novel long noncoding RNA, LINC00483 promotes proliferation and metastasis via modulating of FMNL2 in CRC. Biochem. Biophys. Res. Commun. 2019, 509, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, D.L.C.; Azzarelli, R.; Oishi, K.; Martynoga, B.; Urbán, N.; Dekkers, D.H.W.; Demmers, J.A.; Guillemot, F. Nipbl Interacts with Zfp609 and the Integrator Complex to Regulate Cortical Neuron Migration. Neuron 2017, 93, 348–361. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Xia, J.; Yang, J.; Shi, Y.; Xia, H.; Xiang, X.; Yu, X. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J. BUON 2018, 23, 1343–1349. [Google Scholar]
- Wang, S.; Xue, X.; Wang, R.; Li, X.; Li, Q.; Wang, Y.; Xie, P.; Kang, Y.; Meng, R.; Feng, X. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag. Res. 2018, 10, 3881–3890. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, Y.; Kong, P.; Han, M.; Li, B. Expression of circZNF609 is down-regulated in colorectal cancer tissue and promotes apoptosis in colorectal cancer cells by upregulating p53. Med. Sci. Monit. 2019, 25, 5977–5985. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Ebbesen, K.K.; Sokol, M.; Jakobsen, T.; Korsgaard, U.; Eriksen, A.C.; Hansen, T.B.; Kjems, J.; Hager, H. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. Circinteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rüdel, S.; Flatley, A.; Weinmann, L.; Kremmer, E.; Meister, G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA 2008, 14, 1244–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cawthorne, C.; Swindell, R.; Stratford, I.J.; Dive, C.; Welman, A. Comparison of doxycycline delivery methods for Tet-Inducible gene expression in a subcutaneous xenograft model. J. Biomol. Tech. 2007, 18, 120–123. [Google Scholar] [PubMed]
- Ho, C.K.; Shuman, S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl. Acad. Sci. USA 2002, 99, 12709–12714. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; MacKowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Aken, B.L.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Fernandez Banet, J.; Billis, K.; García Girón, C.; Hourlier, T.; et al. The Ensembl gene annotation system. Database 2016, 2016, baw093. [Google Scholar] [CrossRef]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. CircBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagnon, K.T.; Li, L.; Janowski, B.a.; Corey, D.R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014, 9, 2045–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pall, G.S.; Hamilton, A.J. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008, 3, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. MicroRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho-Xuan, H.; Lehmann, G.; Glazar, P.; Gypas, F.; Eichner, N.; Heizler, K.; Schlitt, H.J.; Zavolan, M.; Rajewsky, N.; Meister, G.; et al. Gene Expression Signatures of a Preclinical Mouse Model during Colorectal Cancer Progression under Low-Dose Metronomic Chemotherapy. Cancers 2021, 13, 49. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13010049
Ho-Xuan H, Lehmann G, Glazar P, Gypas F, Eichner N, Heizler K, Schlitt HJ, Zavolan M, Rajewsky N, Meister G, et al. Gene Expression Signatures of a Preclinical Mouse Model during Colorectal Cancer Progression under Low-Dose Metronomic Chemotherapy. Cancers. 2021; 13(1):49. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13010049
Chicago/Turabian StyleHo-Xuan, Hung, Gerhard Lehmann, Petar Glazar, Foivos Gypas, Norbert Eichner, Kevin Heizler, Hans J. Schlitt, Mihaela Zavolan, Nikolaus Rajewsky, Gunter Meister, and et al. 2021. "Gene Expression Signatures of a Preclinical Mouse Model during Colorectal Cancer Progression under Low-Dose Metronomic Chemotherapy" Cancers 13, no. 1: 49. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13010049