AKR1B1 and AKR1B10 as Prognostic Biomarkers of Endometrioid Endometrial Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Groups
2.2. Immunohistochemistry
2.3. Statistics
2.4. Ethical Issues
3. Results
3.1. Demographic and Histopathological Characteristics of Patients
3.2. AKR1B1 and AKR1B10 Expression Levels in Endometrioid and Serous EC
3.3. Comparison of AKR1B1 and AKR1B10 Expression Levels in Endometrioid and Serous EC and Adjacent Non-Neoplastic Endometrial Tissue
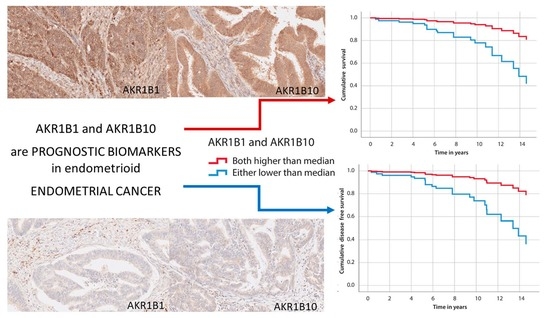
3.4. The Correlation of AKR1B1 and AKR1B10 Expression Levels with Survival
3.5. The Correlation of AKR1B1 and AKR1B10 Expression Levels with Other Clinical Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murali, R.; Delair, D.F.; Bean, S.M.; Abu-Rustum, N.R.; Soslow, R.A. Evolving Roles of Histologic Evaluation and Molecular/Genomic Profiling in the Management of Endometrial Cancer. J. Natl. Compr. Cancer Netw. 2018, 16, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Moore, K.; Brewer, M.A. Endometrial Cancer: Is This a New Disease? Am. Soc. Clin. Oncol. Educ. Book 2017, 37, 435–442. [Google Scholar] [CrossRef]
- Shaw, E.; Farris, M.; McNeil, J.; Friedenreich, C. Obesity and Endometrial Cancer. Recent Results Cancer Res. 2016, 208, 107–136. [Google Scholar] [CrossRef]
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Malik, T.Y.; Chishti, U.; Aziz, A.B.; Sheikh, I. Comparison of Risk Factors and survival of Type 1 and Type II Endometrial Cancers. Pak. J. Med. Sci. 2016, 32, 886–890. [Google Scholar] [CrossRef]
- Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; Benz, C.C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Wang, X.; Jiang, J.; Chen, L. Sentinel lymph node mapping in high-risk endometrial cancer: A systematic review and meta-analysis. Gland. Surg. 2020, 9, 2091–2105. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.C.; Kowalski, L.D.; Scalici, J.; Cantrell, L.; Schuler, K.; Hanna, R.K.; Method, M.; Ade, M.; Ivanova, A.; Boggess, J.F. A comparison of sentinel lymph node biopsy to lymphadenectomy for endometrial cancer staging (FIRES trial): A multicentre, prospective, cohort study. Lancet Oncol. 2017, 18, 384–392. [Google Scholar] [CrossRef]
- Obermair, A.; Abu-Rustum, N.R. Sentinel lymph node mapping in endometrial cancer—areas where further research is needed. Int. J. Gynecol. Cancer 2020, 30, 283–284. [Google Scholar] [CrossRef] [Green Version]
- Schwab, A.; Siddiqui, A.; Vazakidou, M.E.; Napoli, F.; Böttcher, M.; Menchicchi, B.; Raza, U.; Saatci, Ö.; Krebs, A.M.; Ferrazzi, F.; et al. Polyol Pathway Links Glucose Metabolism to the Aggressiveness of Cancer Cells. Cancer Res. 2018, 78, 1604–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khayami, R.; Hashemi, S.R.; Kerachian, M.A. Role of aldo-keto reductase family 1 member B1 (AKR1B1) in the cancer process and its therapeutic potential. J. Cell Mol. Med. 2020, 24, 8890–8902. [Google Scholar] [CrossRef]
- Alzamil, H.A.; Pawade, J.; Fortier, M.A.; Bernal, A.L. Expression of the prostaglandin F synthase AKR1B1 and the prostaglandin transporter SLCO2A1 in human fetal membranes in relation to spontaneous term and preterm labor. Front. Physiol. 2014, 5, 272. [Google Scholar] [CrossRef] [Green Version]
- Kang, E.S.; Kim, G.H.; Kim, H.J.; Woo, I.S.; Ham, S.A.; Jin, H.; Kim, M.Y.; Lee, J.H.; Chang, K.C.; Seo, H.G.; et al. Nrf2 regulates curcumin-induced aldose reductase expression indirectly via nuclear factor-kappaB. Pharmacol. Res. 2008, 58, 15–21. [Google Scholar] [CrossRef]
- Gallego, O.; Ruiz, F.X.; Ardèvol, A.; Domínguez, M.; Alvarez, R.; de Lera, A.R.; Rovira, C.; Farrés, J.; Fita, I.; Parés, X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc. Natl. Acad. Sci. USA 2007, 104, 20764–20769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, T.; Wada, Y.; Endo, S.; Soda, M.; El-Kabbani, O.; Hara, A. Aldo-Keto Reductase 1B10 and Its Role in Proliferation Capacity of Drug-Resistant Cancers. Front. Pharmacol. 2012, 3, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Yan, R.; Zu, X.; Cheng, J.M.; Rao, K.; Liao, D.F.; Cao, D. Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. J. Biol. Chem. 2008, 283, 3418–3423. [Google Scholar] [CrossRef] [Green Version]
- Mounier, C.; Bouraoui, L.; Rassart, E. Lipogenesis in cancer progression (review). Int. J. Oncol. 2014, 45, 485–492. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Shen, H.; Huang, C.; Jing, H.; Cao, D. AKR1B10 induces cell resistance to daunorubicin and idarubicin by reducing C13 ketonic group. Toxicol. Appl. Pharmacol. 2011, 255, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Matsunaga, T.; Suzuki, A.; Kezuka, C.; Okumura, N.; Iguchi, K.; Inoue, I.; Soda, M.; Endo, S.; El-Kabbani, O.; Hara, A.; et al. Aldo-keto reductase 1B10 promotes development of cisplatin resistance in gastrointestinal cancer cells through down-regulating peroxisome proliferator-activated receptor-γ-dependent mechanism. Chem. Biol. Interact. 2016, 256, 142–153. [Google Scholar] [CrossRef]
- Martin, H.J.; Breyer-Pfaff, U.; Wsol, V.; Venz, S.; Block, S.; Maser, E. Purification and characterization of akr1b10 from human liver: Role in carbonyl reduction of xenobiotics. Drug Metab. Dispos. 2006, 34, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Hevir, N.; Sinkovec, J.; Lanišnik Rižner, T. Decreased levels of AKR1B1 and AKR1B10 in cancerous endometrium compared to adjacent non-cancerous tissue. Chem. Biol. Interact. 2013, 202, 226–233. [Google Scholar] [CrossRef]
- Sinreih, M.; Štupar, S.; Čemažar, L.; Verdenik, I.; Frković Grazio, S.; Smrkolj, Š.; Rižner, T.L. STAR and AKR1B10 are down-regulated in high-grade endometrial cancer. J. Steroid Biochem. Mol. Biol. 2017, 171, 43–53. [Google Scholar] [CrossRef]
- Uzozie, A.C.; Selevsek, N.; Wahlander, A.; Nanni, P.; Grossmann, J.; Weber, A.; Buffoli, F.; Marra, G. Targeted Proteomics for Multiplexed Verification of Markers of Colorectal Tumorigenesis. Mol. Cell Proteom. 2017, 16, 407–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzozie, A.; Nanni, P.; Staiano, T.; Grossmann, J.; Barkow-Oesterreicher, S.; Shay, J.W.; Tiwari, A.; Buffoli, F.; Laczko, E.; Marra, G. Sorbitol dehydrogenase overexpression and other aspects of dysregulated protein expression in human precancerous colorectal neoplasms: A quantitative proteomics study. Mol. Cell Proteom. 2014, 13, 1198–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropotova, E.S.; Tychko, R.A.; Zinov’eva, O.L.; Zyrianova, A.F.; Khankin, S.L.; Cherkes, V.L.; Aliev, V.A.; Beresten, S.F.; Oparina, N.I.U.; Mashkova, T.D. Downregulation of AKR1B10 gene expression in colorectal cancer. Mol. Biol. 2010, 44, 243–250. [Google Scholar] [CrossRef]
- Reddy, K.A.; Kumar, P.U.; Srinivasulu, M.; Triveni, B.; Sharada, K.; Ismail, A.; Reddy, G.B. Overexpression and enhanced specific activity of aldoketo reductases (AKR1B1 & AKR1B10) in human breast cancers. Breast 2017, 31, 137–143. [Google Scholar] [CrossRef]
- Knific, T.; Osredkar, J.; Smrkolj, Š.; Tonin, I.; Vouk, K.; Blejec, A.; Frković Grazio, S.; Rižner, T.L. Novel algorithm including CA-125, HE4 and body mass index in the diagnosis of endometrial cancer. Gynecol. Oncol. 2017, 147, 126–132. [Google Scholar] [CrossRef]
- Sinreih, M.; Knific, T.; Anko, M.; Hevir, N.; Vouk, K.; Jerin, A.; Frković Grazio, S.; Rižner, T.L. The Significance of the Sulfatase Pathway for Local Estrogen Formation in Endometrial Cancer. Front. Pharmacol. 2017, 8, 368. [Google Scholar] [CrossRef] [Green Version]
- Penning, T.M. The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef] [Green Version]
- Rižner, T.L. Enzymes of the AKR1B and AKR1C Subfamilies and Uterine Diseases. Front. Pharmacol. 2012, 3, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hojnik, M.; Kenda Šuster, N.; Smrkolj, Š.; Frković Grazio, S.; Verdenik, I.; Rižner, T.L. AKR1C3 Is Associated with Better Survival of Patients with Endometrial Carcinomas. J. Clin. Med. 2020, 9, 4105. [Google Scholar] [CrossRef]
- Taskoparan, B.; Seza, E.G.; Demirkol, S.; Tuncer, S.; Stefek, M.; Gure, A.O.; Banerjee, S. Opposing roles of the aldo-keto reductases AKR1B1 and AKR1B10 in colorectal cancer. Cell Oncol. 2017, 40, 563–578. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.Y.; Lin, Y.H.; Chen, C.L. Overexpression of AKR1B10 predicts tumor recurrence and short survival in oral squamous cell carcinoma patients. J. Oral Pathol. Med. 2019, 48, 712–719. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Jiang, Z.N.; Zheng, Z.H.; Li, Y.; Wang, X.J.; Tang, X. AKR1B10 expression predicts response of gastric cancer to neoadjuvant chemotherapy. Oncol. Lett. 2019, 17, 773–780. [Google Scholar] [CrossRef] [Green Version]
- DiStefano, J.K.; Davis, B. Diagnostic and Prognostic Potential of AKR1B10 in Human Hepatocellular Carcinoma. Cancers 2019, 11, 486. [Google Scholar] [CrossRef] [Green Version]
- Saraswat, M.; Mrudula, T.; Kumar, P.U.; Suneetha, A.; Rao Rao, T.S.; Srinivasulu, M.; Reddy, B. Overexpression of aldose reductase in human cancer tissues. Med. Sci. Monit. 2006, 12, CR525–CR529. [Google Scholar] [PubMed]
- Liu, J.; Zhong, X.; Li, J.; Liu, B.; Guo, S.; Chen, J.; Tan, Q.; Wang, Q.; Ma, W.; Wu, Z.; et al. Screening and identification of lung cancer metastasis-related genes by suppression subtractive hybridization. Thorac. Cancer 2012, 3, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Torres-Mena, J.E.; Salazar-Villegas, K.N.; Sánchez-Rodríguez, R.; López-Gabiño, B.; Del Pozo-Yauner, L.; Arellanes-Robledo, J.; Villa-Treviño, S.; Gutiérrez-Nava, M.A.; Pérez-Carreón, J.I. Aldo-Keto Reductases as Early Biomarkers of Hepatocellular Carcinoma: A Comparison Between Animal Models and Human HCC. Dig. Dis. Sci. 2018, 63, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Demirkol Canlı, S.; Seza, E.G.; Sheraj, I.; Gömçeli, I.; Turhan, N.; Carberry, S.; Prehn, J.H.M.; Güre, A.O.; Banerjee, S. Evaluation of an aldo-keto reductase gene signature with prognostic significance in colon cancer via activation of epithelial to mesenchymal transition and the p70S6K pathway. Carcinogenesis 2020, 41, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.J.; Yeh, Y.C.; Hsu, W.H. Prognostic significance of AKR1B10 in patients with resected lung adenocarcinoma. Thorac. Cancer 2018, 9, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.H.; Cheng, S.L.; Lee, J.J.; Chen, H.M.; Kuo, M.Y.; Cheng, S.J. Expression of AKR1B10 as an independent marker for poor prognosis in human oral squamous cell carcinoma. Head Neck 2017, 39, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, X.; Zhou, D.; Li, H.; Qian, H.; Zhang, J.; Jiang, L.; Wang, B.; Lin, Q.; Zhu, X. Loss of AKR1B10 promotes colorectal cancer cells proliferation and migration via regulating FGF1-dependent pathway. Aging 2020, 12, 13059–13075. [Google Scholar] [CrossRef]
- Yoshitake, H.; Takahashi, M.; Ishikawa, H.; Nojima, M.; Iwanari, H.; Watanabe, A.; Aburatani, H.; Yoshida, K.; Ishi, K.; Takamori, K.; et al. Aldo-keto reductase family 1, member B10 in uterine carcinomas: A potential risk factor of recurrence after surgical therapy in cervical cancer. Int. J. Gynecol. Cancer 2007, 17, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kapoor, A.; Bhatnagar, A. Oxidative and reductive metabolism of lipid-peroxidation derived carbonyls. Chem. Biol. Interact. 2015, 234, 261–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.A.; Choi, B.H.; Nam, C.W.; Song, M.; Kim, S.T.; Lee, J.Y.; Kwak, M.K. Identification of aldo-keto reductases as NRF2-target marker genes in human cells. Toxicol. Lett. 2013, 218, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.M. Aldo-Keto Reductase Regulation by the Nrf2 System: Implications for Stress Response, Chemotherapy Drug Resistance, and Carcinogenesis. Chem. Res. Toxicol. 2017, 30, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.; Blair, I.A.; Van Duyne, G.; Hilser, V.J.; Moiseenkova-Bell, V.; Plymate, S.; Sprenger, C.; Wand, A.J.; Penning, T.M. Using Biochemistry & Biophysics to Extinguish Androgen Receptor Signaling in Prostate Cancer. J. Biol. Chem. 2021, 100240. [Google Scholar] [CrossRef]
- Shen, Y.; Zhong, L.; Johnson, S.; Cao, D. Human aldo-keto reductases 1B1 and 1B10: A comparative study on their enzyme activity toward electrophilic carbonyl compounds. Chem. Biol. Interact. 2011, 191, 192–198. [Google Scholar] [CrossRef] [Green Version]








| Characteristic | Detail | Datum |
|---|---|---|
| Age (y) (n = 113) | Mean ± SD | 63.6 ± 10.1 |
| Weight (kg) (n = 108) a | Mean ± SD | 82.4 ± 17.2 |
| Height (cm) (n = 104) a | Mean ± SD | 162.0 ± 5.5 |
| Body mass index (kg/m2) (n = 104) a | Mean ± SD | 31.5 ± 6.7 |
| Menopausal status (n = 108) (n (%)) a | Postmenopausal | 94 (87.0) |
| Parity status (n = 106) (n (%)) a | Multiparous | 93 (87.7) |
| Smoking status (n = 59) (n (%)) a | Smokers | 5 (8.5) |
| Histological type (n = 113) (n (%)) | Endometrioid | 101 (84.6) |
| Serous | 12 (9.8) | |
| Histological grade (n = 101) (n (%)) | G1 | 65 (62.5) |
| G2 | 25 (24.0) | |
| G3 | 11 (10.6) | |
| Myometrial invasion (n = 113) (n (%)) | <50% | 84 (74.3) |
| ≥50% | 29 (25.7) | |
| Lymphovascular invasion (n = 113) (n (%)) | 30 (26.5) | |
| FIGO stage (n = 108) (n (%)) a | I–II | 97 (89.8) |
| III–IV | 11 (10.1) | |
| Surgical resection (n = 113) (n (%)) | R0 | 108 (95.6) |
| Lymphadenectomy (n = 113) (n (%)) | 105 (92.9) | |
| Adjuvant chemotherapy (n = 113) (n (%)) | Paclitaxel/Carboplatin | 7 (6.2) |
| Carboplatin | 1 (0.1) | |
| Adjuvant radiotherapy (n = 113) (n (%)) | 36 (31.9) | |
| Overall survival (n = 113) (y) | ||
| Range | 0.4–17.6 | |
| Median | 7.6 | |
| Disease-free survival (n = 113) (y) | ||
| Range | 0.2–17.6 | |
| Median | 6.95 |
| Antibody Information | |||||
|---|---|---|---|---|---|
| Antibody | Manufacturer, Catalogue Number, Lot Number | Peptide/Protein Target | Antigen Sequence | Species Raised, Monoclonal, Polyclonal | Dilution |
| Anti-AKR1B1 | Abcam, Cambridge, UK, ab62795,GR64780-2 | Aldo-keto reductase family 1 member B1 | aa 300 to the C-terminus (conjugated to keyhole limpet hemocyanin) | Polyclonal rabbit antibody | 1:200 |
| Anti-AKR1B10 | Abcam, Cambridge, UK, ab96417,GR13314-31 | Aldo-keto reductase family 1 member B10 | fragment corresponding to aa 1-286 | Polyclonal rabbit antibody | 1:200 |
| Antibody Validation | |||||
| Published validation by our research team [22] | |||||
| Current Validation Positive controls for AKR1B1: Kupffer cells, lymphocytes, high-grade serous ovarian cancer cells Positive controls for AKR1B10: Hepatocytes, ductal liver epithelium, lymphocytes, high-grade serous ovarian cancer cells Negative controls for AKR1B1: hepatocytes, fibrous tissue Negative controls for AKR1B10: fibrous tissue | |||||
| Overall Survival | Significance | Hazard Ratio | Confidence Interval |
|---|---|---|---|
| Lymphovascular invasion | p = 0.001 | 3.8 | 1.8–8.2 |
| Risk group (EC grade I–II vs. EC III or SC) | p = 0.005 | 2.9 | 1.4–6.2 |
| AKR1B1 and AKR1B10 expression above the median values | p = 0.036 | 0.4 | 0.1–0.9 |
| FIGO (I–II vs. III–IV) | p = 0.103 | 2.2 | 0.9–5.5 |
| Disease-free survival | |||
| Lymphovascular invasion | p = 0.003 | 3.2 | 1.5–6.7 |
| Risk group (EC grade I–II vs. EC III or SC) | p = 0.004 | 2.9 | 1.4–6.1 |
| AKR1B1 and AKR1B10 expression above the median values | p = 0.023 | 0.3 | 0.1–0.9 |
| FIGO (I–II vs. III–IV) | p = 0.126 | 2.0 | 0.8–5.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hojnik, M.; Frković Grazio, S.; Verdenik, I.; Rižner, T.L. AKR1B1 and AKR1B10 as Prognostic Biomarkers of Endometrioid Endometrial Carcinomas. Cancers 2021, 13, 3398. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13143398
Hojnik M, Frković Grazio S, Verdenik I, Rižner TL. AKR1B1 and AKR1B10 as Prognostic Biomarkers of Endometrioid Endometrial Carcinomas. Cancers. 2021; 13(14):3398. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13143398
Chicago/Turabian StyleHojnik, Marko, Snježana Frković Grazio, Ivan Verdenik, and Tea Lanišnik Rižner. 2021. "AKR1B1 and AKR1B10 as Prognostic Biomarkers of Endometrioid Endometrial Carcinomas" Cancers 13, no. 14: 3398. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13143398