Artificial Intelligence in Thyroid Field—A Comprehensive Review
Abstract
:Simple Summary
Abstract
1. Introduction
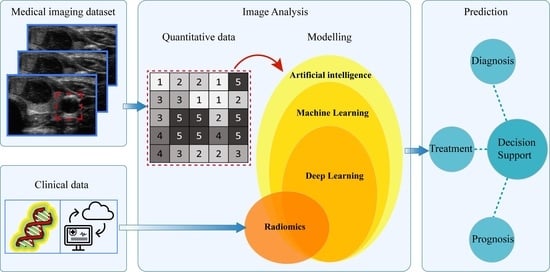
2. Artificial Intelligence in Medical Imaging
2.1. Machine Learning
2.2. Deep Learning
3. Radiomics
- Morphological, that are based on the geometric properties of the ROI, e.g.: volume, maximum surface area, maximum diameter.
- First-order statistics or histogram based, which describe, through histograms, the distribution of grayscale intensity without concern for spatial relationships within the ROI. For instance, calculated features are grey level mean, maximum, minimum and percentiles.
- Second-order statistics or textural features, that represent statistical relationship between the intensity levels of neighboring pixels within the ROI that allow to quantify image heterogeneity, e.g., absolute gradient, grey level co-occurrence matrix (GLCM) grey level run-length matrix (GLRLM), grey level size zone matrix (GLSZM) and grey level distance zone matrix (GLDZM). For instance, GLCM indicates the number of times the same combination of intensity occurs in two pixels separated by a specific distance δ in a known direction.
- Higher-order statistics features, which are computed after the application of mathematical transformation and filters that lead to highlighting repeated patterns, histogram-oriented patterns or local binary patterns, e.g., wavelet or Fourier transforms.
4. AI and Radiomics in Thyroid Diseases
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Machine Learning in Radiation Oncology; El Naqa, I.; Li, R.; Murphy, M. (Eds.) Springer: Cham, Switzerland, 2015. [Google Scholar] [CrossRef]
- Lohmann, P.; Bousabarah, K.; Hoevels, M.; Treuer, H. Radiomics in radiation oncology—basics, methods, and limitations. Strahlenther. Onkol. 2020, 196, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Frix, A.-N.; Cousin, F.; Refaee, T.; Bottari, F.; Vaidyanathan, A.; Desir, C.; Vos, W.; Walsh, S.; Occhipinti, M.; Lovinfosse, P.; et al. Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians. Pers. Med. 2021, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R.; et al. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: Looking into the future. Cancer Cell Int. 2021, 21, 1–11. [Google Scholar] [CrossRef]
- Liang, X.; Yu, J.; Liao, J.; Chen, Z. Convolutional Neural Network for Breast and Thyroid Nodules Diagnosis in Ultrasound Imaging. BioMed Res. Int. 2020, 2020, 1763803. [Google Scholar] [CrossRef]
- Chang, Y.; Paul, A.K.; Kim, N.; Baek, J.H.; Choi, Y.J.; Ha, E.J.; Lee, K.D.; Lee, H.S.; Shin, D.; Kim, N. Computer-aided diagnosis for classifying benign versus malignant thyroid nodules based on ultrasound images: A comparison with radiologist-based assessments. Med. Phys. 2016, 43, 554–567. [Google Scholar] [CrossRef]
- Jin, Z.; Zhu, Y.; Zhang, S.; Xie, F.; Zhang, M.; Zhang, Y.; Tian, X.; Zhang, J.; Luo, Y.; Cao, J. Ultrasound Computer-Aided Diagnosis (CAD) Based on the Thyroid Imaging Reporting and Data System (TI-RADS) to Distinguish Benign from Malignant Thyroid Nodules and the Diagnostic Performance of Radiologists with Different Diagnostic Experience. Med. Sci. Monit. 2020, 26, e918452. [Google Scholar] [CrossRef]
- Fujita, H. AI-based computer-aided diagnosis (AI-CAD): The latest review to read first. Radiol. Phys. Technol. 2020, 13, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, J.J.; Minsky, M.L.; Rochester, N. Artificial Intelligence. Research Laboratory of Electronics (RLE) at the Massachusetts Institute of Technology (MIT). 1959. Available online: https://dspace.mit.edu/handle/1721.1/52263 (accessed on 3 March 2010).
- McCarthy, J.; Minsky, M.L.; Rochester, N.; Shannon, C.E. A proposal for the Dartmouth summer research project on artificial intelligence, August 31, 1955. AI Mag. 2006, 27, 12. [Google Scholar]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Wernick, M.N.; Yang, Y.; Brankov, J.G.; Yourganov, G.; Strother, S. Machine Learning in Medical Imaging. IEEE Signal Process. Mag. 2010, 27, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. RadioGraphics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Zhao, C.-K.; Ren, T.-T.; Yin, Y.-F.; Shi, H.; Wang, H.-X.; Zhou, B.-Y.; Wang, X.-R.; Li, X.; Zhang, Y.-F.; Liu, C.; et al. A Comparative Analysis of Two Machine Learning-Based Diagnostic Patterns with Thyroid Imaging Reporting and Data System for Thyroid Nodules: Diagnostic Performance and Unnecessary Biopsy Rate. Thyroid 2021, 31, 470–481. [Google Scholar] [CrossRef]
- Park, V.; Han, K.; Seong, Y.K.; Park, M.H.; Kim, E.-K.; Moon, H.J.; Yoon, J.H.; Kwak, J.Y. Diagnosis of Thyroid Nodules: Performance of a Deep Learning Convolutional Neural Network Model vs. Radiologists. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Cui, S.; Tseng, H.; Pakela, J.; Haken, R.K.T.; El Naqa, I. Introduction to machine and deep learning for medical physicists. Med. Phys. 2020, 47, e127–e147. [Google Scholar] [CrossRef]
- Forghani, R.; Savadjiev, P.; Chatterjee, A.; Muthukrishnan, N.; Reinhold, C.; Forghani, B. Radiomics and Artificial Intelligence for Biomarker and Prediction Model Development in Oncology. Comput. Struct. Biotechnol. J. 2019, 17, 995–1008. [Google Scholar] [CrossRef] [PubMed]
- Guorong, W.; Dinggang, S.; Mert, R.S. Machine Learning and Medical Imaging; Academic Press: London, UK, 2016. [Google Scholar] [CrossRef]
- El-Naqa, I.; Yang, Y.; Wernick, M.N.; Galatsanos, N.P.; Nishikawa, R. A support vector machine approach for detection of microcalcifications. IEEE Trans. Med. Imaging 2002, 21, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. RadioGraphics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Buda, M.; Saha, A.; Bashir, M.R. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J. Magn. Reson. Imaging 2019, 49, 939–954. [Google Scholar] [CrossRef]
- Wang, L.; Yang, S.; Yang, S.; Zhao, C.; Tian, G.; Gao, Y.; Chen, Y.; Lu, Y. Automatic thyroid nodule recognition and diagnosis in ultrasound imaging with the YOLOv2 neural network. World J. Surg. Oncol. 2019, 17, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Wang, Y.; Yang, X.; Lei, B.; Liu, L.; Li, S.X.; Ni, D.; Wang, T. Deep Learning in Medical Ultrasound Analysis: A Review. Engineering 2019, 5, 261–275. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Kline, T.L.; Akkus, Z.; Philbrick, K.; Weston, A.D. Deep Learning in Radiology: Does One Size Fit All? J. Am. Coll. Radiol. 2018, 15, 521–526. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Avanzo, M.; Stancanello, J.; El Naqa, I. Beyond imaging: The promise of radiomics. Phys. Med. 2017, 38, 122–139. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.-H.; Wei, L.; Cui, S.; Luo, Y.; Haken, R.K.T.; El Naqa, I. Machine Learning and Imaging Informatics in Oncology. Oncology 2020, 98, 344–362. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Jin, Y.; Dai, L.; Zhang, M.; Qiu, Y.; Wang, K.; Tian, J.; Zheng, J. Differential Diagnosis of Benign and Malignant Thyroid Nodules Using Deep Learning Radiomics of Thyroid Ultrasound Images. Eur J. Radiol. 2020, 127, 108992. [Google Scholar] [CrossRef]
- Yoo, Y.J.; Ha, E.J.; Cho, Y.J.; Kim, H.L.; Han, M.; Kang, S.Y. Computer-Aided Diagnosis of Thyroid Nodules via Ultrasonography: Initial Clinical Experience. Korean J. Radiol. 2018, 19, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Agyekum, E.A.; Ren, Y.; Zhang, J.; Zhang, Q.; Sun, H.; Zhang, G.; Xu, F.; Bo, X.; Lv, W.; et al. A Radiomic Nomogram for the Ultrasound-Based Evaluation of Extrathyroidal Extension in Papillary Thyroid Carcinoma. Front. Oncol. 2021, 11, 625646. [Google Scholar] [CrossRef]
- Gillies, R.J.; Schabath, M.B. Radiomics Improves Cancer Screening and Early Detection. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2556–2567. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Tunali, I.; Gillies, R.J.; Schabath, M.B. Application of Radiomics and Artificial Intelligence for Lung Cancer Precision Medicine. Cold Spring Harb. Perspect. Med. 2021, 11, a039537. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhong, X.; Diao, W.; Mu, J.; Cheng, Y.; Jia, Z. Radiomics in Differentiated Thyroid Cancer and Nodules: Explorations; Application; and Limitations. Cancers 2021, 13, 2436. [Google Scholar] [CrossRef] [PubMed]
- Araneo, R.; Bini, F.; Rinaldi, A.; Notargiacomo, A.; Pea, M.; Celozzi, S. Thermal-electric model for piezoelectric ZnO nanowires. Nanotechnology 2015, 26, 265402. [Google Scholar] [CrossRef] [PubMed]
- Scorza, A.; Lupi, G.; Sciuto, S.A.; Bini, F.; Marinozzi, F. A novel approach to a phantom based method for maximum depth of penetration measurement in diagnostic ultrasound: A preliminary study. In Proceedings of the 2015 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Turin, Italy, 7–9 May 2015; pp. 369–374. [Google Scholar] [CrossRef]
- Marinozzi, F.; Branca, F.P.; Bini, F.; Scorza, A. Calibration procedure for performance evaluation of clinical Pulsed Doppler Systems. Measurement 2012, 45, 1334–1342. [Google Scholar] [CrossRef]
- Shen, Y.-T.; Chen, L.; Yue, W.-W.; Xu, H.-X. Artificial intelligence in ultrasound. Eur. J. Radiol. 2021, 139. [Google Scholar] [CrossRef]
- Zhang, B.; Tian, J.; Pei, S.; Chen, Y.; He, X.; Dong, Y.; Zhang, L.; Mo, X.; Huang, W.; Cong, S.; et al. Machine Learning-Assisted System for Thyroid Nodule Diagnosis. Thyroid 2019, 29, 858–867. [Google Scholar] [CrossRef]
- Wu, G.G.; Lv, W.Z.; Yin, R.; Xu, J.W.; Yan, Y.J.; Chen, R.X.; Wang, J.Y.; Zhang, B.; Cui, X.W.; Dietrich, C.F. Deep Learning Based on ACR TI-RADS Can Improve the Differential Diagnosis of Thyroid Nodules. Front. Oncol. 2021, 11, 575166. [Google Scholar] [CrossRef]
- Koh, J.; Lee, E.; Han, K.; Kim, E.-K.; Son, E.J.; Sohn, Y.-M.; Seo, M.; Kwon, M.-R.; Yoon, J.H.; Lee, J.H.; et al. Diagnosis of thyroid nodules on ultrasonography by a deep convolutional neural network. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Ko, S.Y.; Lee, J.H.; Yoon, J.H.; Na, H.; Hong, E.; Han, K.; Jung, I.; Kim, E.K.; Moon, H.J.; Park, V.Y.; et al. Deep convolutional neural network for the diagnosis of thyroid nodules on ultrasound. Head Neck 2019, 41, 885–891. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Zhang, Q.; Wei, X.; Pan, Y.; Zhao, J.; Xin, X.; Qin, C.; Wang, X.; Li, J.; et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: A retrospective; multicohort; diagnostic study. Lancet Oncol. 2019, 20, 193–201. [Google Scholar] [CrossRef]
- Ma, J.; Wu, F.; Zhu, J.; Xu, D.; Kong, D. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics 2017, 73, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Buda, M.; Wildman-Tobriner, B.; Hoang, J.K.; Thayer, D.; Tessler, F.N.; Middleton, W.D.; Mazurowski, M.A. Management of Thyroid Nodules Seen on US Images: Deep Learning May Match Performance of Radiologists. Radiology 2019, 292, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Chi, J.; Walia, E.; Babyn, P.; Wang, J.; Groot, G.; Eramian, M. Thyroid Nodule Classification in Ultrasound Images by Fine-Tuning Deep Convolutional Neural Network. J. Digit. Imaging. 2017, 30, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.R.; Lee, E.; Kim, H.R.; Yoon, J.H.; Park, V.Y.; Kwak, J.Y. Convolutional Neural Network to Stratify the Malignancy Risk of Thyroid Nodules: Diagnostic Performance Compared with the American College of Radiology Thyroid Imaging Reporting and Data System Implemented by Experienced Radiologists. AJNR Am. J. Neuroradiol. 2021, 42, 1513–1519. [Google Scholar] [CrossRef] [PubMed]
- Park, V.Y.; Lee, E.; Lee, H.S.; Kim, H.J.; Yoon, J.; Son, J.; Song, K.; Moon, H.J.; Yoon, J.H.; Kim, G.R.; et al. Combining radiomics with ultrasound-based risk stratification systems for thyroid nodules: An approach for improving performance. Eur. Radiol. 2021, 31, 2405–2413. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Wang, H.; Wang, L.; Hu, W.; Sun, X.; Dai, Z.; Zhu, J.; Li, H.; Ge, Y.; Song, B. Radiomics based on multiparametric MRI for extrathyroidal extension feature prediction in papillary thyroid cancer. BMC Med. Imaging 2021, 21, 20. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.-R.; Shin, J.; Park, H.; Cho, H.; Hahn, S.; Park, K. Radiomics Study of Thyroid Ultrasound for Predicting BRAF Mutation in Papillary Thyroid Carcinoma: Preliminary Results. Am. J. Neuroradiol. 2020, 41, 700–705. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Zhu, J.; Qiu, Q.; Wang, Y.; Bai, T.; Yin, Y. Prediction of Immunohistochemistry of Suspected Thyroid Nodules by Use of Machine Learning-Based Radiomics. AJR Am. J. Roentgenol. 2019, 213, 1348–1357. [Google Scholar] [CrossRef]
- Guo, R.; Guo, J.; Zhang, L.; Qu, X.; Dai, S.; Peng, R.; Chong, V.F.H.; Xian, J. CT-based radiomics features in the prediction of thyroid cartilage invasion from laryngeal and hypopharyngeal squamous cell carcinoma. Cancer Imaging 2020, 20, 81. [Google Scholar] [CrossRef]
- Park, V.; Han, K.; Lee, E.; Kim, E.-K.; Moon, H.J.; Yoon, J.H.; Kwak, J.Y. Association Between Radiomics Signature and Disease-Free Survival in Conventional Papillary Thyroid Carcinoma. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yue, W.; Li, X.; Liu, S.; Guo, L.; Xu, H.; Zhang, H.; Yang, G. Comparison Study of Radiomics and Deep Learning-Based Methods for Thyroid Nodules Classification Using Ultrasound Images. IEEE Access 2020, 8, 52010–52017. [Google Scholar] [CrossRef]
- Peng, S.; Liu, Y.; Lv, W.; Liu, L.; Zhou, Q.; Yang, H.; Ren, J.; Liu, G.; Wang, X.; Zhang, X.; et al. Deep learning-based artificial intelligence model to assist thyroid nodule diagnosis and management: A multicentre diagnostic study. Lancet Digit. Health 2021, 3, e250–e259. [Google Scholar] [CrossRef]
- Trimboli, P.; Bini, F.; Andrioli, M.; Giovanella, L.; Thorel, M.F.; Ceriani, L.; Valabrega, S.; Lenzi, A.; Drudi, F.M.; Marinozzi, F.; et al. Analysis of tissue surrounding thyroid nodules by ultrasound digital images. Endocrine 2015, 48, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.; Wu, K.; Daneshjou, R.; Ouyang, D.; Ho, D.E.; Zou, J. How medical AI devices are evaluated: Limitations and recommendations from an analysis of FDA approvals. Nat. Med. 2021, 27, 582–584. [Google Scholar] [CrossRef] [PubMed]
- Verburg, F.; Reiners, C. Sonographic diagnosis of thyroid cancer with support of AI. Nat. Rev. Endocrinol. 2019, 15, 319–321. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Bini, F.; Trimboli, P.; Marinozzi, F.; Giovanella, L. Treatment of benign thyroid nodules by high intensity focused ultrasound (HIFU) at different acoustic powers: A study on in-silico phantom. Endocrine 2018, 59, 506–509. [Google Scholar] [CrossRef]
- Trimboli, P.; Bini, F.; Baek, J.H.; Marinozzi, F.; Giovanella, L. High intensity focused ultrasounds (HIFU) therapy for benign thyroid nodules without anesthesia or sedation. Endocrine 2018, 61, 210–215. [Google Scholar] [CrossRef]
- Giovanella, L.; Piccardo, A.; Pezzoli, C.; Bini, F.; Ricci, R.; Ruberto, T.; Trimboli, P. Comparison of High Intensity Focused Ultrasound and radioiodine for treating toxic Thyroid nodules. Clin. Endocrinol. 2018, 89, 219–225. [Google Scholar] [CrossRef]
- Benjamens, S.; Dhunnoo, P.; Mesko, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef]
- Ben-Israel, D.; Jacobs, W.B.; Casha, S.; Lang, S.; Ryu, W.H.A.; de Lotbiniere-Bassett, M.; Cadotte, D.W. The impact of machine learning on patient care: A systematic review. Artif. Intell. Med. 2020, 103, 101785. [Google Scholar] [CrossRef]
- Russ, G.; Trimboli, P.; Buffet, C. The New Era of TIRADSs to Stratify the Risk of Malignancy of Thyroid Nodules: Strengths, Weaknesses and Pitfalls. Cancers 2021, 13, 4316. [Google Scholar] [CrossRef] [PubMed]
- Trimboli, P. Ultrasound: The Extension of Our Hands to Improve the Management of Thyroid Patients. Cancers 2021, 13, 567. [Google Scholar] [CrossRef] [PubMed]
- Scappaticcio, L.; Piccardo, A.; Treglia, G.; Poller, D.N.; Trimboli, P. The dilemma of 18F-FDG PET/CT thyroid incidentaloma: What we should expect from FNA. A systematic review and meta-analysis. Endocrine 2021, 73, 540–549. [Google Scholar] [CrossRef] [PubMed]




| Study | Description | Cohort | Method | Performance |
|---|---|---|---|---|
| Zhao et al., 2021 [21] | Classification | 106 patients | SVM | Accuracy: 82% |
| Benign/malignant thyroid nodules | Sensitivity: 91% | |||
| US | Specificity: 78% | |||
| Park et al., 2019 [22] | Classification | 286 patients | SVM | Accuracy: 75.9% |
| Benign/malignant thyroid nodules | Sensitivity: 90.4% | |||
| US | Specificity: 58.8% | |||
| Zhang et al., 2019 [51] | Classification | 826 patients | SVM | Accuracy: 83% |
| Benign/malignant thyroid nodules | Sensitivity: 86.1% | |||
| US | Specificity: 82.7% | |||
| Yoo et al., 2018 [41] | Classification | 50 patients | SVM | Accuracy: 84.6% |
| Benign/malignant thyroid nodules | Sensitivity: 80% | |||
| US | Specificity: 88.1% | |||
| Chang et al., 2016 [10] | Classification | 118 patients | SVM | Accuracy: 98.3% |
| Benign/malignant thyroid nodules | Sensitivity: N/A | |||
| US | Specificity: N/A |
| Study | Description | Cohort | Method | Performance |
|---|---|---|---|---|
| Kim et al., 2021 [59] | Malignancy risk thyroid modules | 757 patients | CNN | Accuracy: 85.1% |
| Sensitivity: 81.8% | ||||
| Specificity: 86.1% | ||||
| Wu et al., 2021 [52] | Classification | 1396 patients | CNN | Accuracy: 82% |
| Benign/malignant thyroid nodules | Sensitivity: 85% | |||
| US | Specificity: 78% | |||
| Jin et al., 2020 [11] | Classification | 695 patients | CNN | Accuracy: 80.3% |
| Benign/malignant thyroid nodules | Sensitivity: 80.6% | |||
| US | Specificity: 80.1% | |||
| Liang et al., 2020 [9] | Classification | 221 patients | CNN | Accuracy: 75% |
| Benign/malignant thyroid nodules | Sensitivity: 84.9% | |||
| US | Specificity: 69% | |||
| Buda et al., 2019 [57] | Nodule detection | 1230 patients | CNN | Accuracy: N/A |
| Predict malignancy | Sensitivity: 87% | |||
| Risk level stratification | Specificity: 52% | |||
| Ko et al., 2019 [54] | Classification | 519 patients | CNN | Accuracy: 87.3% |
| Benign/malignant thyroid nodules | Sensitivity: 90% | |||
| US | Specificity: 82% | |||
| Park et al., 2019 [22] | Classification | 286 patients | CNN | Accuracy: 86% |
| Benign/malignant thyroid nodules | Sensitivity:91% | |||
| US | Specificity: 80% | |||
| Wang et al., 2019 [33] | Classification | 276 patients | CNN | Accuracy: 90.3% |
| Benign/malignant thyroid nodules | Sensitivity: 90.5% | |||
| US | Specificity: 89.91% | |||
| Li et al., 2018 [55] | Classification | 17 627 patients | CNN | Accuracy: 86% |
| Benign/malignant thyroid nodules | Sensitivity: 84% | |||
| US | Specificity: 87% | |||
| Chi et al., 2017 [58] | Classification | 592 patients | CNN | Accuracy: 96.3% |
| Benign/malignant thyroid nodules | Sensitivity: 82.8% | |||
| US | Specificity: 99.3% | |||
| Ma et al., 2017 [56] | Classification | 4782 patients | CNN | Accuracy: 83% |
| Benign/malignant thyroid nodules | Sensitivity: 82.4% | |||
| US | Specificity: 84.9% |
| Study | Description | Cohort | Method | Performance |
|---|---|---|---|---|
| Park et al., 2021 [60] | Classification: Benign/malignant thyroid nodules 730 features extracted and 66 selected US | 1609 patients | ML-based radiomics | Accuracy: 77.8% |
| Sensitivity: 70.6% | ||||
| Specificity: 79.8% | ||||
| Peng et al., 2021 [67] | Classification Benign/malignant thyroid nodules US | 8339 patients | DL-based radiomics | Accuracy: 89.1% |
| Sensitivity: 94.9% | ||||
| Specificity: 81.2% | ||||
| Wang et al., 2021 [42] | Evaluation of extrathyroidal extension (ETE) in patients with papillary thyroid carcinoma; 479 features extracted; 10 features selected US | 132 patients | ML-based radiomics | Accuracy: 83% |
| Sensitivity: 65% | ||||
| Specificity: 74% | ||||
| Wei et al., 2021 [61] | Evaluation of extrathyroidal extension (ETE) in patients with papillary thyroid carcinoma MRI | 102 patients | ML-based radiomics | Accuracy: 79% |
| Sensitivity: 75% | ||||
| Specificity: 80% | ||||
| Zhao et al., 2021 [21] | Classification | 106 patients | ML-based radiomics | Accuracy: 75.5% |
| Benign/malignant thyroid nodules | Sensitivity: 69.7% | |||
| US | Specificity: 78.1% | |||
| Guo et al., 2020 [64] | Prediction of thyroid cartilage invasion from Laryngeal and hypopharyngeal squamous cell carcinoma; 1029 features extracted; 30 features selected CT images | 265 patients | ML-based radiomics | Accuracy: 90% |
| Sensitivity: 80.2% | ||||
| Specificity: 88.3% | ||||
| Kwon et al., 2020 [62] | Predict the presence or absence of BRAF proto-oncogene, serine/threonine kinase (BRAF) mutation in papillary thyroid cancer US | 96 patients | ML-based radiomics | Accuracy: 64.3% |
| Sensitivity: 66.8% | ||||
| Specificity: 61.8% | ||||
| Wang et al., 2020 [66] | Classification | 1040 patients | ML-based radiomics | Accuracy: 66.8% |
| Benign/malignant thyroid nodules | Sensitivity: 51.2% | |||
| US | Specificity: 75.8% | |||
| Zhou et al., 2020 [40] | Classification | 1734 patients | DL-based radiomics | Accuracy: 97% |
| Benign/malignant thyroid nodules | Sensitivity: 89.5% | |||
| US | Specificity: 84.1% | |||
| Gu et al., 2019 [63] | Evaluating immunohistochemical characteristics in patients with suspected thyroid nodules CT images | 103 patients | ML-based radiomics | Accuracy: 84% |
| Sensitivity: 93% | ||||
| Specificity: 73% | ||||
| Park et al., 2019 [65] | Estimate disease free survival rate in patients with papillary thyroid carcinoma; | 768 patients | ML-based radiomics | Accuracy: 77% |
| 730 features extracted and 40 selected | Sensitivity: N/A | |||
| US | Specificity: N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bini, F.; Pica, A.; Azzimonti, L.; Giusti, A.; Ruinelli, L.; Marinozzi, F.; Trimboli, P. Artificial Intelligence in Thyroid Field—A Comprehensive Review. Cancers 2021, 13, 4740. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13194740
Bini F, Pica A, Azzimonti L, Giusti A, Ruinelli L, Marinozzi F, Trimboli P. Artificial Intelligence in Thyroid Field—A Comprehensive Review. Cancers. 2021; 13(19):4740. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13194740
Chicago/Turabian StyleBini, Fabiano, Andrada Pica, Laura Azzimonti, Alessandro Giusti, Lorenzo Ruinelli, Franco Marinozzi, and Pierpaolo Trimboli. 2021. "Artificial Intelligence in Thyroid Field—A Comprehensive Review" Cancers 13, no. 19: 4740. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13194740