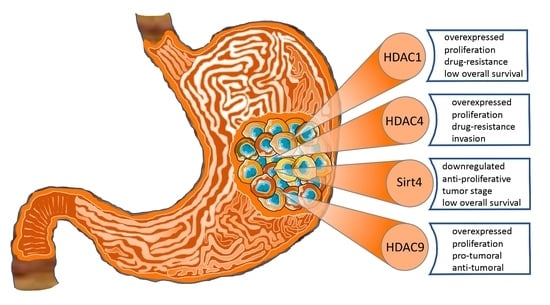
Histone Deacetylase Functions in Gastric Cancer: Therapeutic Target?
Abstract
:Simple Summary
Abstract
1. Introduction
2. Histone Deacetylases
2.1. Generalities
2.2. In Gastric Cancer (GC)
2.2.1. HDACs of Class I
2.2.2. HDACs of Class IIa
2.2.3. HDACs of Class IIb
2.2.4. HDACs of Class III
2.2.5. HDAC of Class IV
2.2.6. Conclusions
3. Histone Deacetylase Inhibitors
3.1. Generalities
3.2. In Gastric Cancer
3.2.1. Belinostat
3.2.2. Panobinostat
3.2.3. Romidepsin
3.2.4. Vorinostat
3.2.5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Eom, S.S.; Choi, W.; Eom, B.W.; Park, S.H.; Kim, S.J.; Kim, Y.I.; Yoon, H.M.; Lee, J.Y.; Kim, C.G.; Kim, H.K.; et al. A Comprehensive and Comparative Review of Global Gastric Cancer Treatment Guidelines. J. Gastric Cancer 2022, 22, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Barsouk, A. Epidemiology of Gastric Cancer: Global Trends, Risk Factors and Prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- American Cancer Society Key Statistics About Stomach Cancer. Available online: https://www.cancer.org/cancer/stomach-cancer/about/key-statistics.html (accessed on 3 June 2020).
- Garcia-Pelaez, J.; Barbosa-Matos, R.; São José, C.; Sousa, S.; Gullo, I.; Hoogerbrugge, N.; Carneiro, F.; Oliveira, C. Gastric Cancer Genetic Predisposition and Clinical Presentations: Established Heritable Causes and Potential Candidate Genes. Eur. J. Med. Genet 2022, 65, 104401. [Google Scholar] [CrossRef]
- Oliveira, C.; Pinheiro, H.; Figueiredo, J.; Seruca, R.; Carneiro, F. Familial Gastric Cancer: Genetic Susceptibility, Pathology, and Implications for Management. Lancet Oncol. 2015, 16, e60–e70. [Google Scholar] [CrossRef]
- Laurén, P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and so-called Intestinal-Type Carcinoma: An Attempt at a Histo-Clinical Classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Petrelli, F.; Berenato, R.; Turati, L.; Mennitto, A.; Steccanella, F.; Caporale, M.; Dallera, P.; de Braud, F.; Pezzica, E.; Di Bartolomeo, M.; et al. Prognostic Value of Diffuse versus Intestinal Histotype in Patients with Gastric Cancer: A Systematic Review and Meta-Analysis. J. Gastrointest. Oncol. 2017, 8, 148–163. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [CrossRef] [Green Version]
- Camargo, M.C.; Kim, W.-H.; Chiaravalli, A.M.; Kim, K.-M.; Corvalan, A.H.; Matsuo, K.; Yu, J.; Sung, J.J.Y.; Herrera-Goepfert, R.; Meneses-Gonzalez, F.; et al. Improved Survival of Gastric Cancer with Tumour Epstein–Barr Virus Positivity: An International Pooled Analysis. Gut 2014, 63, 236–243. [Google Scholar] [CrossRef]
- Sohn, B.H.; Hwang, J.-E.; Jang, H.-J.; Lee, H.-S.; Oh, S.C.; Shim, J.-J.; Lee, K.-W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef]
- Zhu, L.; Lu, Z.; Wang, Y.; Zhang, C.; Liu, Y.; Qu, X. Microsatellite Instability and Survival in Gastric Cancer: A Systematic Review and Meta-Analysis. Mol. Clin. Oncol. 2015, 3, 699–705. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular Analysis of Gastric Cancer Identifies Subtypes Associated with Distinct Clinical Outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Mottamal, M.; Zheng, S.; Huang, T.L.; Wang, G. Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef] [Green Version]
- Gigek, C.O.; Chen, E.S.; Calcagno, D.Q.; Wisnieski, F.; Burbano, R.R.; Smith, M.A.C. Epigenetic Mechanisms in Gastric Cancer. Epigenomics 2012, 4, 279–294. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [Green Version]
- Bondarev, A.D.; Attwood, M.M.; Jonsson, J.; Chubarev, V.N.; Tarasov, V.V.; Schiöth, H.B. Recent Developments of HDAC Inhibitors: Emerging Indications and Novel Molecules. Br. J. Clin. Pharmacol. 2021, 87, 4577–4597. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [Green Version]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and Deacetylation of Non-Histone Proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef]
- Hassell, K.N. Histone Deacetylases and Their Inhibitors in Cancer Epigenetics. Diseases 2019, 7, 57. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Kwon, H.J.; Yoon, B.; Kim, J.; Han, S.U.; Joo, H.J.; Kim, D. Expression Profile of Histone Deacetylase 1 in Gastric Cancer Tissues. Jpn. J. Cancer Res. 2001, 92, 1300–1304. [Google Scholar] [CrossRef]
- Deng, R.; Zhang, P.; Liu, W.; Zeng, X.; Ma, X.; Shi, L.; Wang, T.; Yin, Y.; Chang, W.; Zhang, P.; et al. HDAC Is Indispensable for IFN-γ-Induced B7-H1 Expression in Gastric Cancer. Clin. Epigenet. 2018, 10, 153. [Google Scholar] [CrossRef]
- Eto, S.; Yoshikawa, K.; Shimada, M.; Higashijima, J.; Tokunaga, T.; Nakao, T.; Nishi, M.; Takasu, C.; Sato, H.; Kurita, N. The Relationship of CD133, Histone Deacetylase 1 and Thrombospondin-1 in Gastric Cancer. Anticancer Res. 2015, 35, 2071–2076. [Google Scholar]
- Gao, F.; Lv, Y.; Zhu, Y.; Chen, M.; Shen, S.; Cao, J.; Zou, X. Correlation of Epigenetic Aberrance with STAT3 Signaling Pathway in Gastric Carcinogenesis. Dig. Dis. Sci. 2012, 57, 2055–2062. [Google Scholar] [CrossRef]
- Jiang, Z.; Sun, X.; Zhang, Q.; Ji, X.; Yu, Q.; Huang, T.; Chen, D.; Chen, H.; Mei, X.; Wang, L.; et al. Identification of Candidate Biomarkers That Involved in the Epigenetic Transcriptional Regulation for Detection Gastric Cancer by ITRAQ Based Quantitative Proteomic Analysis. Clin. Chim. Acta 2017, 471, 29–37. [Google Scholar] [CrossRef]
- Jiang, Z.; Yang, H.; Zhang, X.; Wang, Z.; Rong, R.; Wang, X. Histone Deacetylase-1 as a Prognostic Factor and Mediator of Gastric Cancer Progression by Enhancing Glycolysis. Hum. Pathol. 2019, 85, 194–201. [Google Scholar] [CrossRef]
- Kim, J.-H.; Choi, Y.-K.; Kwon, H.-J.; Yang, H.-K.; Choi, J.-H.; Kim, D.-Y. Downregulation of Gelsolin and Retinoic Acid Receptor β Expression in Gastric Cancer Tissues through Histone Deacetylase 1. J. Gastroenterol. Hepatol. 2004, 19, 218–224. [Google Scholar] [CrossRef]
- Sudo, T.; Mimori, K.; Nishida, N.; Kogo, R.; Iwaya, T.; Tanaka, F.; Shibata, K.; Fujita, H.; Shirouzu, K.; Mori, M. Histone Deacetylase 1 Expression in Gastric Cancer. Oncol. Rep. 2011, 26, 777–782. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Piao, J.; Li, N.; Yang, Y.; Kim, K.-Y.; Lin, Z. Valproic Acid Targets HDAC1/2 and HDAC1/PTEN/Akt Signalling to Inhibit Cell Proliferation via the Induction of Autophagy in Gastric Cancer. FEBS J. 2020, 287, 2118–2133. [Google Scholar] [CrossRef]
- Yu, Z.; Zeng, J.; Liu, H.; Wang, T.; Yu, Z.; Chen, J. Role of HDAC1 in the Progression of Gastric Cancer and the Correlation with LncRNAs. Oncol. Lett. 2019, 17, 3296–3304. [Google Scholar] [CrossRef] [Green Version]
- Wisnieski, F.; Calcagno, D.Q.; Leal, M.F.; Chen, E.S.; Gigek, C.O.; Santos, L.C.; Pontes, T.B.; Rasmussen, L.T.; Payão, S.L.M.; Assumpção, P.P.; et al. Differential Expression of Histone Deacetylase and Acetyltransferase Genes in Gastric Cancer and Their Modulation by Trichostatin A. Tumour Biol. 2014, 35, 6373–6381. [Google Scholar] [CrossRef]
- He, Q.; Li, G.; Wang, X.; Wang, S.; Hu, J.; Yang, L.; He, Y.; Pan, Y.; Yu, D.; Wu, Y. A Decrease of Histone Deacetylase 6 Expression Caused by Helicobacter Pylori Infection Is Associated with Oncogenic Transformation in Gastric Cancer. Cell. Physiol. Biochem. 2017, 42, 1326–1335. [Google Scholar] [CrossRef]
- Mutze, K.; Langer, R.; Becker, K.; Ott, K.; Novotny, A.; Luber, B.; Hapfelmeier, A.; Göttlicher, M.; Höfler, H.; Keller, G. Histone Deacetylase (HDAC) 1 and 2 Expression and Chemotherapy in Gastric Cancer. Ann. Surg. Oncol. 2010, 17, 3336–3343. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, Y.; Chen, W.; Wang, X.; Miao, Z.; Cao, L.; Li, F.; Wang, G. By Recruiting HDAC1, MORC2 Suppresses P21 Waf1/Cip1 in Gastric Cancer. Oncotarget 2015, 6, 16461–16470. [Google Scholar] [CrossRef] [Green Version]
- Xia, G.; Schneider-Stock, R.; Diestel, A.; Habold, C.; Krueger, S.; Roessner, A.; Naumann, M.; Lendeckel, U. Helicobacter Pylori Regulates P21WAF1 by Histone H4 Acetylation. Biochem. Biophys. Res. Commun. 2008, 369, 526–531. [Google Scholar] [CrossRef]
- Pero, R.; Peluso, S.; Angrisano, T.; Tuccillo, C.; Sacchetti, S.; Keller, S.; Tomaiuolo, R.; Bruni, C.B.; Lembo, F.; Chiariotti, L. Chromatin and DNA Methylation Dynamics of Helicobacter Pylori-Induced COX-2 Activation. Int. J. Med. Microbiol. 2011, 301, 140–149. [Google Scholar] [CrossRef]
- Shen, Q.; Tang, W.; Sun, J.; Feng, L.; Jin, H.; Wang, X. Regulation of CRADD-Caspase 2 Cascade by Histone Deacetylase 1 in Gastric Cancer. Am. J. Transl. Res. 2014, 6, 538–547. [Google Scholar]
- Regel, I.; Merkl, L.; Friedrich, T.; Burgermeister, E.; Zimmermann, W.; Einwächter, H.; Herrmann, K.; Langer, R.; Röcken, C.; Hofheinz, R.; et al. Pan-Histone Deacetylase Inhibitor Panobinostat Sensitizes Gastric Cancer Cells to Anthracyclines via Induction of CITED2. Gastroenterology 2012, 143, 99–109.e10. [Google Scholar] [CrossRef]
- Wu, S.; Wu, E.; Wang, D.; Niu, Y.; Yue, H.; Zhang, D.; Luo, J.; Chen, R. LncRNA HRCEG, Regulated by HDAC1, Inhibits Cells Proliferation and Epithelial-Mesenchymal-Transition in Gastric Cancer. Cancer Genet. 2020, 241, 25–33. [Google Scholar] [CrossRef]
- Lin, L.; Jiang, H.; Huang, M.; Hou, X.; Sun, X.; Jiang, X.; Dong, X.; Sun, X.; Zhou, B.; Qiao, H. Depletion of Histone Deacetylase 1 Inhibits Metastatic Abilities of Gastric Cancer Cells by Regulating the MiR-34a/CD44 Pathway. Oncol. Rep. 2015, 34, 663–672. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.; Yao, Q.; Sun, J.; Feng, L.; Lu, H.; Ma, Y.; Liu, L.; Wang, F.; Li, J.; Yue, Y.; et al. Downregulation of Histone Deacetylase 1 by MicroRNA-520h Contributes to the Chemotherapeutic Effect of Doxorubicin. FEBS Lett. 2014, 588, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Noh, J.H.; Eun, J.W.; Jung, K.H.; Bae, H.J.; Shen, Q.; Kim, M.G.; Chang, Y.G.; Kim, S.-J.; Park, W.S.; et al. Targeted Inactivation of HDAC2 Restores P16INK4a Activity and Exerts Antitumor Effects on Human Gastric Cancer. Mol. Cancer Res. 2013, 11, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Orenay-Boyacioglu, S.; Kasap, E.; Gerceker, E.; Yuceyar, H.; Demirci, U.; Bilgic, F.; Korkmaz, M. Expression Profiles of Histone Modification Genes in Gastric Cancer Progression. Mol. Biol. Rep. 2018, 45, 2275–2282. [Google Scholar] [CrossRef]
- Song, J.; Noh, J.H.; Lee, J.H.; Eun, J.W.; Ahn, Y.M.; Kim, S.Y.; Lee, S.H.; Park, W.S.; Yoo, N.J.; Lee, J.Y.; et al. Increased Expression of Histone Deacetylase 2 Is Found in Human Gastric Cancer. APMIS 2005, 113, 264–268. [Google Scholar] [CrossRef]
- Wei, J.; Wang, Z.; Wang, Z.; Yang, Y.; Fu, C.; Zhu, J.; Jiang, D. MicroRNA-31 Function as a Suppressor Was Regulated by Epigenetic Mechanisms in Gastric Cancer. Biomed. Res. Int. 2017, 2017, 5348490. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Kang, W.; Lu, X.; Ma, S.; Dong, L.; Zou, B. Weighted Gene Co-Expression Network Analysis and Connectivity Map Identifies Lovastatin as a Treatment Option of Gastric Cancer by Inhibiting HDAC2. Gene 2019, 681, 15–25. [Google Scholar] [CrossRef]
- Nakagawa, M.; Oda, Y.; Eguchi, T.; Aishima, S.-I.; Yao, T.; Hosoi, F.; Basaki, Y.; Ono, M.; Kuwano, M.; Tanaka, M.; et al. Expression Profile of Class I Histone Deacetylases in Human Cancer Tissues. Oncol. Rep. 2007, 18, 769–774. [Google Scholar] [CrossRef]
- Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Ebert, M.P.; Pross, M.; Dietel, M.; Denkert, C.; Röcken, C. Association of Patterns of Class I Histone Deacetylase Expression with Patient Prognosis in Gastric Cancer: A Retrospective Analysis. Lancet Oncol. 2008, 9, 139–148. [Google Scholar] [CrossRef]
- Xu, G.; Zhu, H.; Zhang, M.; Xu, J. Histone Deacetylase 3 Is Associated with Gastric Cancer Cell Growth via the MiR-454-Mediated Targeting of CHD5. Int. J. Mol. Med. 2018, 41, 155–163. [Google Scholar] [CrossRef]
- Yang, Z.; Jiang, X.; Zhang, Z.; Zhao, Z.; Xing, W.; Liu, Y.; Jiang, X.; Zhao, H. HDAC3-Dependent Transcriptional Repression of FOXA2 Regulates FTO/M6A/MYC Signaling to Contribute to the Development of Gastric Cancer. Cancer Gene 2021, 28, 141–155. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, F.; Meng, Z.; Luo, Q.; Pan, D.; Qian, Y. Inhibited HDAC3 Promotes MicroRNA-376c-3p to Suppress Malignant Phenotypes of Gastric Cancer Cells by Reducing WNT2b. Genomics 2021, 113, 3512–3522. [Google Scholar] [CrossRef]
- Wu, S.-M.; Jan, Y.-J.; Tsai, S.-C.; Pan, H.-C.; Shen, C.-C.; Yang, C.-N.; Lee, S.-H.; Liu, S.-H.; Shen, L.-W.; Chiu, C.-S.; et al. Targeting Histone Deacetylase-3 Blocked Epithelial-Mesenchymal Plasticity and Metastatic Dissemination in Gastric Cancer. Cell Biol. Toxicol. 2022, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, Y.; Xu, P.; Yang, R.; Ma, Z.; Liang, S.; Zhang, G. The Inhibition of Histone Deacetylase 8 Suppresses Proliferation and Inhibits Apoptosis in Gastric Adenocarcinoma. Int. J. Oncol. 2015, 47, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Colarossi, L.; Memeo, L.; Colarossi, C.; Aiello, E.; Iuppa, A.; Espina, V.; Liotta, L.; Mueller, C. Inhibition of Histone Deacetylase 4 Increases Cytotoxicity of Docetaxel in Gastric Cancer Cells. Proteom.–Clin. Appl. 2014, 8, 924–931. [Google Scholar] [CrossRef]
- Kang, Z.-H.; Wang, C.-Y.; Zhang, W.-L.; Zhang, J.-T.; Yuan, C.-H.; Zhao, P.-W.; Lin, Y.-Y.; Hong, S.; Li, C.-Y.; Wang, L. Histone Deacetylase HDAC4 Promotes Gastric Cancer SGC-7901 Cells Progression via P21 Repression. PLoS ONE 2014, 9, e98894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, K.; Wei, Y.; Yao, Q.; Zhang, Q.; Qu, H.; Zhu, G. LncRNA-MIAT Regulates Cell Biological Behaviors in Gastric Cancer through a Mechanism Involving the MiR-29a-3p/HDAC4 Axis. Oncol. Rep. 2017, 38, 3465–3472. [Google Scholar] [CrossRef] [Green Version]
- Spaety, M.-E.; Gries, A.; Badie, A.; Venkatasamy, A.; Romain, B.; Orvain, C.; Yanagihara, K.; Okamoto, K.; Jung, A.C.; Mellitzer, G.; et al. HDAC4 Levels Control Sensibility toward Cisplatin in Gastric Cancer via the P53-P73/BIK Pathway. Cancers 2019, 11, 1747. [Google Scholar] [CrossRef] [Green Version]
- Zang, W.-J.; Hu, Y.-L.; Qian, C.-Y.; Feng, Y.; Liu, J.-Z.; Yang, J.-L.; Huang, H.; Zhu, Y.-Z.; Xue, W.-J. HDAC4 Promotes the Growth and Metastasis of Gastric Cancer via Autophagic Degradation of MEKK3. Br. J. Cancer 2022, 127, 237–248. [Google Scholar] [CrossRef]
- Zhang, E.; He, X.; Zhang, C.; Su, J.; Lu, X.; Si, X.; Chen, J.; Yin, D.; Han, L.; De, W. A Novel Long Noncoding RNA HOXC-AS3 Mediates Tumorigenesis of Gastric Cancer by Binding to YBX1. Genome Biol. 2018, 19, 154. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Zhao, J.; Yang, Z.; Zhou, Z.; Lu, P. Identification of Gene Signatures and Potential Therapeutic Targets for Acquired Chemotherapy Resistance in Gastric Cancer Patients. J. Gastrointest. Oncol. 2021, 12, 407–422. [Google Scholar] [CrossRef]
- Li, A.; Liu, Z.; Li, M.; Zhou, S.; Xu, Y.; Xiao, Y.; Yang, W. HDAC5, a Potential Therapeutic Target and Prognostic Biomarker, Promotes Proliferation, Invasion and Migration in Human Breast Cancer. Oncotarget 2016, 7, 37966–37978. [Google Scholar] [CrossRef]
- Yu, Y.; Cao, F.; Yu, X.; Zhou, P.; Di, Q.; Lei, J.; Tai, Y.; Wu, H.; Li, X.; Wang, X.; et al. The Expression of HDAC7 in Cancerous Gastric Tissues Is Positively Associated with Distant Metastasis and Poor Patient Prognosis. Clin. Transl. Oncol. 2017, 19, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, L.; Yuan, C.; Wang, C.; Gao, T.; Zheng, Z. MiR-489 Inhibited the Development of Gastric Cancer via Regulating HDAC7 and PI3K/AKT Pathway. World J. Surg. Oncol. 2020, 18, 73. [Google Scholar] [CrossRef] [PubMed]
- Xiong, K.; Zhang, H.; Du, Y.; Tian, J.; Ding, S. Identification of HDAC9 as a Viable Therapeutic Target for the Treatment of Gastric Cancer. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Li, N.; Zhang, Y.; Zhang, J.; Xu, R.; Wu, Y. MicroRNA-383-5p Inhibits the Progression of Gastric Carcinoma via Targeting HDAC9 Expression. Braz. J. Med. Biol. Res. 2019, 52. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Chen, J.; Shi, Y.; Shen, R.; Chen, M. Expression of HDAC9 in Gastric. Int. J. Clin. Exp. Pathol. 2016, 9, 12829–12835. [Google Scholar]
- Park, S.J.; Kim, J.K.; Bae, H.J.; Eun, J.W.; Shen, Q.; Kim, H.S.; Shin, W.C.; Yang, H.D.; Lee, E.K.; You, J.S.; et al. HDAC6 Sustains Growth Stimulation by Prolonging the Activation of EGF Receptor through the Inhibition of Rabaptin-5-Mediated Early Endosome Fusion in Gastric Cancer. Cancer Lett. 2014, 354, 97–106. [Google Scholar] [CrossRef]
- Jin, Z.; Jiang, W.; Jiao, F.; Guo, Z.; Hu, H.; Wang, L.; Wang, L. Decreased Expression of Histone Deacetylase 10 Predicts Poor Prognosis of Gastric Cancer Patients. Int. J. Clin. Exp. Pathol. 2014, 7, 5872–5879. [Google Scholar]
- Cha, E.J.; Noh, S.J.; Kwon, K.S.; Kim, C.Y.; Park, B.-H.; Park, H.S.; Lee, H.; Chung, M.J.; Kang, M.J.; Lee, D.G.; et al. Expression of DBC1 and SIRT1 Is Associated with Poor Prognosis of Gastric Carcinoma. Clin. Cancer Res. 2009, 15, 4453–4459. [Google Scholar] [CrossRef] [Green Version]
- Feng, A.N.; Zhang, L.H.; Fan, X.S.; Huang, Q.; Ye, Q.; Wu, H.Y.; Yang, J. Expression of SIRT1 in Gastric Cardiac Cancer and Its Clinicopathologic Significance. Int. J. Surg. Pathol. 2011, 19, 743–750. [Google Scholar] [CrossRef]
- Kang, Y.; Jung, W.Y.; Lee, H.; Lee, E.; Kim, A.; Kim, B.-H. Expression of SIRT1 and DBC1 in Gastric Adenocarcinoma. Korean J. Pathol. 2012, 46, 523–531. [Google Scholar] [CrossRef]
- Noguchi, A.; Kikuchi, K.; Zheng, H.; Takahashi, H.; Miyagi, Y.; Aoki, I.; Takano, Y. SIRT1 Expression Is Associated with a Poor Prognosis, Whereas DBC1 Is Associated with Favorable Outcomes in Gastric Cancer. Cancer Med. 2014, 3, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Özcan, Ö.; Belli, A.K.; Çetin, E.S.; Kara, M.; Çelik, Ö.İ.; Kaplan, M.; Kayılıoğlu, S.I.; Dönmez, C.; Polat, M. Upregulation of SIRT1 Gene in Gastric Adenocarcinoma. Turk. J. Gastroenterol. 2019, 30, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Li, X.; Wei, C.; Che, X.; He, S.; Lu, J.; Wang, S.; Pang, K.; Fan, L. The Prognostic Role of SIRT1-Autophagy Axis in Gastric Cancer. Dis. Mrk. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wang, X.; Chen, P. MiR-204 down Regulates SIRT1 and Reverts SIRT1-Induced Epithelial-Mesenchymal Transition, Anoikis Resistance and Invasion in Gastric Cancer Cells. BMC Cancer 2013, 13, 290. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, P.; Huang, Z.; Hu, X.; Chen, M.; Hu, S.; Hu, Y.; Cai, T. Sirt7 Promotes Gastric Cancer Growth and Inhibits Apoptosis by Epigenetically Inhibiting MiR-34a. Sci. Rep. 2015, 5, 9787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Huang, S.; Deng, C.; Cao, Y.; Yang, J.; Chen, G.; Zhang, B.; Duan, C.; Shi, J.; Kong, B.; et al. Co-Ordinated Overexpression of SIRT1 and STAT3 Is Associated with Poor Survival Outcome in Gastric Cancer Patients. Oncotarget 2017, 8, 18848–18860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Yang, Y.; Huang, S.; Deng, C.; Zhou, S.; Yang, J.; Cao, Y.; Xu, L.; Yuan, Y.; Yang, J.; et al. SIRT1 Inhibits Gastric Cancer Proliferation and Metastasis via STAT3/MMP-13 Signaling. J. Cell. Physiol. 2019, 234, 15395–15406. [Google Scholar] [CrossRef]
- An, Y.; Wang, B.; Wang, X.; Dong, G.; Jia, J.; Yang, Q. SIRT1 Inhibits Chemoresistance and Cancer Stemness of Gastric Cancer by Initiating an AMPK/FOXO3 Positive Feedback Loop. Cell Death Dis. 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Wang, B.; Gao, W.; Huang, S.; Liu, Z.; Li, W.; Jia, J. SIRT1 Is Downregulated in Gastric Cancer and Leads to G1-Phase Arrest via NF-ΚB/Cyclin D1 Signaling. Mol. Cancer Res. 2013, 11, 1497–1507. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S.; Zhang, L. Upregulated MiR-132 in Lgr5+ Gastric Cancer Stem Cell-like Cells Contributes to Cisplatin-Resistance via SIRT1/CREB/ABCG2 Signaling Pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, X.; Chai, N.; Gu, Y.; Zhang, S.; Ding, M.; Cao, H.; Sha, S.; Yin, J.; Li, M.; et al. SIRT1 Is Reduced in Gastric Adenocarcinoma and Acts as a Potential Tumor Suppressor in Gastric Cancer. Gastrointest. Tumors 2015, 2, 109–123. [Google Scholar] [CrossRef]
- Szász, A.M.; Lánczky, A.; Nagy, Á.; Förster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabó, A.; Győrffy, B. Cross-Validation of Survival Associated Biomarkers in Gastric Cancer Using Transcriptomic Data of 1,065 Patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Qiu, C.; Sun, W.; Gu, M.; Xiao, F.; Zou, J.; Zhang, L. Yap Regulates Gastric Cancer Survival and Migration via SIRT1/Mfn2/Mitophagy. Oncol. Rep. 2018, 39, 1671–1681. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Liao, K.; Liu, D. MiRNA-12129 Suppresses Cell Proliferation and Block Cell Cycle Progression by Targeting SIRT1 in GASTRIC Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820928144. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Ko, Y.S.; Park, J.; Choi, Y.; Park, J.-W.; Kim, Y.; Pyo, J.-S.; Yoo, Y.B.; Lee, J.-S.; Lee, B.L. Forkhead Transcription Factor FOXO1 Inhibits Angiogenesis in Gastric Cancer in Relation to SIRT1. Cancer Res. Treat. 2016, 48, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Dong, G.; Wang, B.; An, Y.; Li, J.; Wang, X.; Jia, J.; Yang, Q. SIRT1 Suppresses the Migration and Invasion of Gastric Cancer by Regulating ARHGAP5 Expression. Cell Death Dis. 2018, 9, 977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Zhang, L.; Chen, X.; Lu, Q.; Yang, Y.; Liu, J.; Ma, X. SIRT1 Counteracted the Activation of STAT3 and NF-ΚB to Repress the Gastric Cancer Growth. Int. J. Clin. Exp. Med. 2014, 7, 5050–5058. [Google Scholar]
- Yang, Q.; Wang, B.; Zang, W.; Wang, X.; Liu, Z.; Li, W.; Jia, J. Resveratrol Inhibits the Growth of Gastric Cancer by Inducing G1 Phase Arrest and Senescence in a Sirt1-Dependent Manner. PLoS ONE 2013, 8, e70627. [Google Scholar] [CrossRef]
- Li, J.; Dong, G.; Wang, B.; Gao, W.; Yang, Q. MiR-543 Promotes Gastric Cancer Cell Proliferation by Targeting SIRT1. Biochem. Biophys. Res. Commun. 2016, 469, 15–21. [Google Scholar] [CrossRef]
- Luo, D.; Fan, H.; Ma, X.; Yang, C.; He, Y.; Ge, Y.; Jiang, M.; Xu, Z.; Yang, L. MiR-1301-3p Promotes Cell Proliferation and Facilitates Cell Cycle Progression via Targeting SIRT1 in Gastric Cancer. Front. Oncol. 2021, 11, 664242. [Google Scholar] [CrossRef]
- Zhu, H.; Xia, L.; Zhang, Y.; Wang, H.; Xu, W.; Hu, H.; Wang, J.; Xin, J.; Gang, Y.; Sha, S.; et al. Activating Transcription Factor 4 Confers a Multidrug Resistance Phenotype to Gastric Cancer Cells through Transactivation of SIRT1 Expression. PLoS ONE 2012, 7, e31431. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-Y.; Cheng, H.-L.; Lee, Y.-H.; Yuan, T.-M.; Chen, S.-W.; Lin, Y.-Y.; Chueh, P.J. Tumor-Associated NADH Oxidase (TNOX)-NAD+-Sirtuin 1 Axis Contributes to Oxaliplatin-Induced Apoptosis of Gastric Cancer Cells. Oncotarget 2017, 8, 15338–15348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, M.; Dorfman, R.G.; Pan, Y.; Tang, D.; Xu, L.; Zhao, Z.; Zhou, Q.; Zhou, L.; Wang, Y.; et al. SIRT2 Promotes the Migration and Invasion of Gastric Cancer through RAS/ERK/JNK/MMP-9 Pathway by Increasing PEPCK1-Related Metabolism. Neoplasia 2018, 20, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, J.; Wang, L.; Min, X.; Chen, Z. The LINC00152/MiR-138 Axis Facilitates Gastric Cancer Progression by Mediating SIRT2. J. Oncol. 2021, 2021, 1173869. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-H.; Hsu, C.-C.; Fang, W.-L.; Chi, C.-W.; Sung, M.-T.; Kao, H.-L.; Li, A.F.-Y.; Yin, P.-H.; Yang, M.-H.; Lee, H.-C. SIRT3 Expression as a Biomarker for Better Prognosis in Gastric Cancer. World J. Surg. 2014, 38, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhao, G.; Du, J.; Li, J.; Lin, G.; Zhang, J. LncRNA FENDRR Inhibits Gastric Cancer Cell Proliferation and Invasion via the MiR-421/SIRT3/Notch-1 Axis. Cancer Manag. Res. 2021, 13, 9175–9187. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.-Y.; Cao, L.-P. SIRT3 Inhibits Cell Proliferation in Human Gastric Cancer through Down-Regulation of Notch-1. Int. J. Clin. Exp. Med. 2015, 8, 5263–5271. [Google Scholar]
- Yang, B.; Fu, X.; Shao, L.; Ding, Y.; Zeng, D. Aberrant Expression of SIRT3 Is Conversely Correlated with the Progression and Prognosis of Human Gastric Cancer. Biochem. Biophys. Res. Commun. 2014, 443, 156–160. [Google Scholar] [CrossRef]
- Cui, Y.; Qin, L.; Wu, J.; Qu, X.; Hou, C.; Sun, W.; Li, S.; Vaughan, A.T.M.; Li, J.J.; Liu, J. SIRT3 Enhances Glycolysis and Proliferation in SIRT3-Expressing Gastric Cancer Cells. PLoS ONE 2015, 10, e0129834. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Jung, D.E.; Yu, S.S.; Lee, Y.S.; Choi, B.K.; Lee, Y.C. Regulation of SIRT3 Signal Related Metabolic Reprogramming in Gastric Cancer by Helicobacter Pylori Oncoprotein CagA. Oncotarget 2017, 8, 78365–78378. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.-Y.; Xu, Q.; Wu, D.-D.; Lau, A.T.Y.; Xu, Y.-M. The Prognostic and Clinicopathological Roles of Sirtuin-3 in Various Cancers. PLoS ONE 2016, 11, e0159801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Cui, F.; Yu, F.; Lu, H.; Zhang, M.; Tang, H.; Peng, Z. Sirtuin-4 (SIRT4) Is Downregulated and Associated with Some Clinicopathological Features in Gastric Adenocarcinoma. Biomed. Pharmacother. 2015, 72, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Huang, D.; Liu, G.; Jian, F.; Zhu, J.; Zhang, L. SIRT4 Acts as a Tumor Suppressor in Gastric Cancer by Inhibiting Cell Proliferation, Migration, and Invasion. Oncol. Targets 2018, 11, 3959–3968. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Lin, J.; Lin, Y.; Chen, X.; Zhu, G.; Huang, G. Overexpression of SIRT4 Inhibits the Proliferation of Gastric Cancer Cells through Cell Cycle Arrest. Oncol. Lett. 2019, 17, 2171–2176. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Qian, Q.; Xu, Y.; Xu, X.; Zhang, L.; He, S.; Li, D. SIRT5 Regulates Autophagy and Apoptosis in Gastric Cancer Cells. J. Int. Med. Res. 2021, 49, 300060520986355. [Google Scholar] [CrossRef]
- Lu, X.; Yang, P.; Zhao, X.; Jiang, M.; Hu, S.; Ouyang, Y.; Zeng, L.; Wu, J. OGDH Mediates the Inhibition of SIRT5 on Cell Proliferation and Migration of Gastric Cancer. Exp. Cell Res. 2019, 382, 111483. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, L.; Tang, Y.; Xie, D.; Wu, K.; Wei, W.; Xiao, Q. CDK2 Positively Regulates Aerobic Glycolysis by Suppressing SIRT5 in Gastric Cancer. Cancer Sci. 2018, 109, 2590–2598. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, C.; Li, X.; Shen, J.; Xu, Y.; Shi, H.; Mu, X.; Pan, J.; Zhao, T.; Li, M.; et al. CPT1A-mediated Succinylation of S100A10 Increases Human Gastric Cancer Invasion. J. Cell Mol. Med. 2019, 23, 293–305. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, A.; Yu, X.; Zhu, J.; Dai, H. SIRT6 Inhibits Growth of Gastric Cancer by Inhibiting JAK2/STAT3 Pathway. Oncol. Rep. 2017, 38, 1059–1066. [Google Scholar] [CrossRef] [Green Version]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA Approval Summary: Vorinostat for Treatment of Advanced Primary Cutaneous T-Cell Lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef]
- Poole, R.M. Belinostat: First Global Approval. Drugs 2014, 74, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Raedler, L.A. Farydak (Panobinostat): First HDAC Inhibitor Approved for Patients with Relapsed Multiple Myeloma. Am. Health Drug Benefits 2016, 9, 84–87. [Google Scholar] [PubMed]
- Iyer, S.P.; Foss, F.F. Romidepsin for the Treatment of Peripheral T-Cell Lymphoma. Oncologist 2015, 20, 1084–1091. [Google Scholar] [CrossRef] [Green Version]
- Ramaiah, M.J.; Tangutur, A.D.; Manyam, R.R. Epigenetic Modulation and Understanding of HDAC Inhibitors in Cancer Therapy. Life Sci. 2021, 277, 119504. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.-R.; Kim, D.-Y.; Jin, H.; Meng, R.; Chai, O.H.; Kim, S.-H.; Park, B.-H.; Kim, S.M. Inactivation of the Akt/FOXM1 Signaling Pathway by Panobinostat Suppresses the Proliferation and Metastasis of Gastric Cancer Cells. Int. J. Mol. Sci. 2021, 22, 5955. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-L.; Chao, Y.-J.; Wang, H.-C.; Hou, Y.-C.; Chen, C.G.; Chang, C.-C.; Shan, Y.-S. Local Ablation of Gastric Cancer by Reconstituted Apolipoprotein B Lipoparticles Carrying Epigenetic Drugs. Nanomed. Nanotechnol. Biol. Med. 2021, 37, 102450. [Google Scholar] [CrossRef]
- Sasaki, Y.; Negishi, H.; Idogawa, M.; Suzuki, H.; Mita, H.; Toyota, M.; Shinomura, Y.; Imai, K.; Tokino, T. Histone Deacetylase Inhibitor FK228 Enhances Adenovirus-Mediated P53 Family Gene Therapy in Cancer Models. Mol. Cancer Ther. 2008, 7, 779–787. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Adachi, M.; Zhao, X.; Kawamura, R.; Imai, K. Histone Deacetylase Inhibitors FK228, N-(2-Aminophenyl)-4-[N-(Pyridin-3-Yl-Methoxycarbonyl)Amino- Methyl]Benzamide and m-Carboxycinnamic Acid Bis-Hydroxamide Augment Radiation-Induced Cell Death in Gastrointestinal Adenocarcinoma Cells. Int. J. Cancer 2004, 110, 301–308. [Google Scholar] [CrossRef]
- Adachi, M.; Zhang, Y.; Zhao, X.; Minami, T.; Kawamura, R.; Hinoda, Y.; Imai, K. Synergistic Effect of Histone Deacetylase Inhibitors FK228 and M-Carboxycinnamic Acid Bis-Hydroxamide with Proteasome Inhibitors PSI and PS-341 against Gastrointestinal Adenocarcinoma Cells. Clin. Cancer Res. 2004, 10, 3853–3862. [Google Scholar] [CrossRef] [Green Version]
- Hui, K.F.; Cheung, A.K.L.; Choi, C.K.; Yeung, P.L.; Middeldorp, J.M.; Lung, M.L.; Tsao, S.W.; Chiang, A.K.S. Inhibition of Class I Histone Deacetylases by Romidepsin Potently Induces Epstein-Barr Virus Lytic Cycle and Mediates Enhanced Cell Death with Ganciclovir. Int. J. Cancer 2016, 138, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Park, J.K.; Seo, J.S.; Lee, S.K.; Chan, K.K.; Kuh, H.-J. Combinatorial Antitumor Activity of Oxaliplatin with Epigenetic Modifying Agents, 5-Aza-CdR and FK228, in Human Gastric Cancer Cells. Biomol. Ther. 2018, 26, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Rha, S.Y.; Jeung, H.-C.; Jung, J.-J.; Kim, T.S.; Kwon, H.J.; Kim, B.S.; Chung, H.C. Identification of Genes Related to a Synergistic Effect of Taxane and Suberoylanilide Hydroxamic Acid Combination Treatment in Gastric Cancer Cells. J. Cancer Res. Clin. Oncol. 2010, 136, 1901–1913. [Google Scholar] [CrossRef] [PubMed]
- Claerhout, S.; Lim, J.Y.; Choi, W.; Park, Y.-Y.; Kim, K.; Kim, S.-B.; Lee, J.-S.; Mills, G.B.; Cho, J.Y. Gene Expression Signature Analysis Identifies Vorinostat as a Candidate Therapy for Gastric Cancer. PLoS ONE 2011, 6, e24662. [Google Scholar] [CrossRef]
- Decourtye-Espiard, L.; Bougen-Zhukov, N.; Godwin, T.; Brew, T.; Schulpen, E.; Black, M.A.; Guilford, P. E-Cadherin-Deficient Epithelial Cells Are Sensitive to HDAC Inhibitors. Cancers 2021, 14, 175. [Google Scholar] [CrossRef]
- Hibino, S.; Saito, Y.; Muramatsu, T.; Otani, A.; Kasai, Y.; Kimura, M.; Saito, H. Inhibitors of Enhancer of Zeste Homolog 2 (EZH2) Activate Tumor-Suppressor MicroRNAs in Human Cancer Cells. Oncogenesis 2014, 3, e104. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ida, H.; Ito, K.; Zhang, H.; Ito, Y. Contribution of Reactivated RUNX3 to Inhibition of Gastric Cancer Cell Growth Following Suberoylanilide Hydroxamic Acid (Vorinostat) Treatment. Biochem. Pharmacol. 2007, 73, 990–1000. [Google Scholar] [CrossRef]
- Lu, H.; Yang, X.-F.; Tian, X.-Q.; Tang, S.-L.; Li, L.-Q.; Zhao, S.; Zheng, H.-C. The in Vitro and Vivo Anti-Tumor Effects and Molecular Mechanisms of Suberoylanilide Hydroxamic Acid (SAHA) and MG132 on the Aggressive Phenotypes of Gastric Cancer Cells. Oncotarget 2016, 7, 56508–56525. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Ji, J.; Shi, M.; Yang, L.; Yu, Y.; Liu, B.; Zhu, Z.; Zhang, J. Suberoylanilide Hydroxamic Acid Enhances the Antitumor Activity of Oxaliplatin by Reversing the Oxaliplatin-induced Src Activation in Gastric Cancer Cells. Mol. Med. Rep. 2014, 10, 2729–2735. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Liu, S.; Zhang, J.; Pei, M.; Xiao, Y.; Li, J.; Hong, L.; Lin, J.; Wang, J.; Wu, X.; et al. Vorinostat Triggers MiR-769–5p/3p-Mediated Suppression of Proliferation and Induces Apoptosis via the STAT3-IGF1R-HDAC3 Complex in Human Gastric Cancer. Cancer Lett. 2021, 521, 196–209. [Google Scholar] [CrossRef]
- Gou, W.; Yang, X.; Shen, D.; Zhao, S.; Liu, Y.; Sun, H.; Takano, Y.; Su, R.; Luo, J.; Zheng, H. The Roles of BTG3 Expression in Gastric Cancer: A Potential Marker for Carcinogenesis and a Target Molecule for Gene Therapy. Oncotarget 2015, 6, 19841–19867. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Li, J.; Shen, D.; Yang, X.; Zhao, S.; Wu, Y.; Takano, Y.; Sun, H.; Su, R.; Luo, J.; et al. BTG1 Expression Correlates with Pathogenesis, Aggressive Behaviors and Prognosis of Gastric Cancer: A Potential Target for Gene Therapy. Oncotarget 2015, 6, 19685–19705. [Google Scholar] [CrossRef] [Green Version]
- Labisso, W.L.; Wirth, M.; Stojanovic, N.; Stauber, R.H.; Schnieke, A.; Schmid, R.M.; Krämer, O.H.; Saur, D.; Schneider, G. MYC Directs Transcription of MCL1 and EIF4E Genes to Control Sensitivity of Gastric Cancer Cells toward HDAC Inhibitors. Cell Cycle 2012, 11, 1593–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Das, K.; Wu, J.; Lee, M.H.; Tan, P. RNH1 Regulation of Reactive Oxygen Species Contributes to Histone Deacetylase Inhibitor Resistance in Gastric Cancer Cells. Oncogene 2014, 33, 1527–1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, W.; Shen, D.; Yang, X.; Zhao, S.; Liu, Y.; Sun, H.; Su, R.-J.; Luo, J.; Zheng, H. ING5 Suppresses Proliferation, Apoptosis, Migration and Invasion, and Induces Autophagy and Differentiation of Gastric Cancer Cells: A Good Marker for Carcinogenesis and Subsequent Progression. Oncotarget 2015, 6, 19552–19579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, K.F.; Chiang, A.K.S. Suberoylanilide Hydroxamic Acid Induces Viral Lytic Cycle in Epstein-Barr Virus-Positive Epithelial Malignancies and Mediates Enhanced Cell Death. Int. J. Cancer 2010, 126, 2479–2489. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Mori, H.; Matsuzaki, J.; Sato, A.; Saito, Y.; Imoto, M.; Suematsu, M.; Suzuki, H. CAPZA1 Determines the Risk of Gastric Carcinogenesis by Inhibiting Helicobacter Pylori CagA-Degraded Autophagy. Autophagy 2019, 15, 242–258. [Google Scholar] [CrossRef]
- Venkatasamy, A.; Guerin, E.; Blanchet, A.; Orvain, C.; Devignot, V.; Jung, M.; Jung, A.C.; Chenard, M.-P.; Romain, B.; Gaiddon, C.; et al. Ultrasound and Transcriptomics Identify a Differential Impact of Cisplatin and Histone Deacetylation on Tumor Structure and Microenvironment in a Patient-Derived In Vivo Model of Gastric Cancer. Pharmaceutics 2021, 13, 1485. [Google Scholar] [CrossRef]
- Seah, K.S.; Loh, J.Y.; Nguyen, T.T.T.; Tan, H.L.; Hutchinson, P.E.; Lim, K.K.; Dymock, B.W.; Long, Y.C.; Lee, E.J.D.; Shen, H.-M.; et al. SAHA and Cisplatin Sensitize Gastric Cancer Cells to Doxorubicin by Induction of DNA Damage, Apoptosis and Perturbation of AMPK-MTOR Signalling. Exp. Cell Res. 2018, 370, 283–291. [Google Scholar] [CrossRef]
- Yoo, C.; Ryu, M.-H.; Na, Y.-S.; Ryoo, B.-Y.; Lee, C.-W.; Maeng, J.; Kim, S.-Y.; Koo, D.H.; Park, I.; Kang, Y.-K. Phase I and Pharmacodynamic Study of Vorinostat Combined with Capecitabine and Cisplatin as First-Line Chemotherapy in Advanced Gastric Cancer. Investig. New Drugs 2014, 32, 271–278. [Google Scholar] [CrossRef]
- Yoo, C.; Ryu, M.-H.; Na, Y.-S.; Ryoo, B.-Y.; Lee, C.-W.; Kang, Y.-K. Vorinostat in Combination with Capecitabine plus Cisplatin as a First-Line Chemotherapy for Patients with Metastatic or Unresectable Gastric Cancer: Phase II Study and Biomarker Analysis. Br. J. Cancer 2016, 114, 1185–1190. [Google Scholar] [CrossRef]

| Family | Class | Member | Yeast Counterpart | Subcellular Localization | |
|---|---|---|---|---|---|
| Zn2+-dependent | I | HDAC 1, 2, 3, 8 | Rpd3 | Nucleus | |
| II | a | HDAC 4, 5, 7, 9 | Hda1 | Nucleus/Cytoplasm | |
| b | HDAC 6, 10 | ||||
| IV | HDAC 11 | Nucleus | |||
| NAD+-dependent | III | SIRT 1, 2 | Sir2 | Nucleus/Cytoplasm | |
| SIRT 3, 4, 5 | Mitochondria | ||||
| SIRT 6, 7 | Nucleus | ||||
| Class | Member | Regulation | Target | Cellular Process Implication | Clinicopathological Implication | Reference |
|---|---|---|---|---|---|---|
| I | HDAC1 | Up, None, or Down* | CITED2 CRADD HIF-1α lncRNA AF116637 lncRNA BC01600 lncRNA HRCEG mir-34a/CD44 p21 | Cell proliferation Apoptosis Glycolysis EMT Chemosensitivity | Age H. pylori infection Lauren classification Lymph node metastasis Lymphovascular invasion Tumor size Tumor stage Survival* | [21,23,24,25,26,27,28,30,32,33,34,35,36,37,38,39,40,41] |
| HDAC2 | Up or None* | CITED2 p16INK4a | Cell proliferation Autophagy Apoptosis Chemosensitivity | Lauren classification* Tumor grade Tumor stage Survival* | [29,33,38,42,44,44,45,46,48] | |
| HDAC3 | Up or None* | FOXA2/FTO/MYC miR-376/WNT2b mir-454/CHD5 | Cell proliferation Apoptosis Cell invasion Cell migration Tumor growth | LNM Tumor grade Tumor infiltration depth Tumor stage Survival | [22,31,47,49,50,51,52] | |
| HDAC8 | Up or None* | Cell cycle arrest Cell proliferation Apoptosis | LNM Tumor grade Tumor stage | [47,53] | ||
| IIa | HDAC4 | Up | ATG4B/MEKK3/p38 BIK MIAT/miR-29a-3p P21 | Cell cycle arrest Cell proliferation Apoptosis Autophagy Cell invasion Cell migration Tumor growth Chemosensitivity | H. pylori infection Lauren’s classification LNM Molecular subgroup Tumor depth invasion Tumor stage Survival | [32,54,55,56,57,58] |
| HDAC5 | Up or Down* | HOXC-AS3/YBX1 | Cell proliferation Cell migration | Survival | [43,59,60,61] | |
| HDAC7 | Up | Cell proliferation Cell migration Cell invasion | Survival | [63] | ||
| HDAC9 | Up or None* | Cell proliferation Apoptosis Tumor growth | Tumor stage* Survival* | [64,65,66] | ||
| IIb | HDAC6 | Up or Down* | EGFR/Rabaptin-5 | Cell proliferation | H. pylori infection Tumor progression Survival | [32,67] |
| HDAC10 | Down | [68] | ||||
| III | SIRT1 | Up or Down* | AMPK/FOXO3c-JUN/ARHGAP5 miR-12129 miR-132/SIRT1/CREB/ABCG2 miR-204/SIRT1/LKB1 NFκB/cyclin D1 STAT3/MMP-13 Yap/SIRT1/Mfn2 | Cell proliferation Apoptosis Autophagy Senescence Cell invasion Cell migration EMT Mitophagy Chemo-sensitivity* | Sex Age Lauren’s classification Tumor grade Tumor size Tumor invasion LNM Tumor stage Survival* | [69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93] |
| III | SIRT2 | Up | PEPCK1 | Cell cycle arrest Cell proliferation Apoptosis Cell invasion Cell migration Tumor growth Metastases | Survival | [94,95] |
| SIRT3 | Up or Down* | HIF-1α LDHA Notch-1 | Cell proliferation Cell invasion ROS homeostasis Tumor growth | Tumor grade Surival | [96,97,98,99,100,101,102] | |
| SIRT4 | Down | E-cadherin | Cell proliferation Cell invasion Cell migration EMT | LNM Tumor stage Survival | [103,104,105] | |
| SIRT5 | Down | OGDH S100A10 | Cell cycle arrest Cell proliferation Cell invasion Cell migration EMT | Lymphovascular invasion Nodal involvement Survival | [106,107,108,109] | |
| SIRT6 | Down | JAK2/STAT3 | Cell cycle arrest Cell proliferation Apoptosis Tumor growth | Tumor grade Tumor size Tumor stage Survival | [110] | |
| SIRT7 | Up | mir-34a | Cell proliferation Apoptosis Tumor growth | Extent of gastrectomy Lymph node status Tumor depth invasion Tumor size Metastases Tumor stage Survival | [76] | |
| IV | HDAC11 | no data available |
| Class | Member | |||
|---|---|---|---|---|
| Diagnostic | Prognostic | Predictive | ||
| I | HDAC1 | |||
| HDAC2 | ||||
| HDAC3 | ||||
| HDAC8 | ||||
| IIa | HDAC4 | |||
| HDAC5 | ||||
| HDAC7 | ||||
| HDAC9 | ||||
| IIb | HDAC6 | |||
| HDAC10 | ||||
| III | SIRT1 | |||
| SIRT2 | ||||
| SIRT3 | ||||
| SIRT4 | ||||
| SIRT5 | ||||
| SIRT6 | ||||
| SIRT7 | ||||
| IV | HDAC11 | no | no | no |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badie, A.; Gaiddon, C.; Mellitzer, G. Histone Deacetylase Functions in Gastric Cancer: Therapeutic Target? Cancers 2022, 14, 5472. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14215472
Badie A, Gaiddon C, Mellitzer G. Histone Deacetylase Functions in Gastric Cancer: Therapeutic Target? Cancers. 2022; 14(21):5472. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14215472
Chicago/Turabian StyleBadie, Amandine, Christian Gaiddon, and Georg Mellitzer. 2022. "Histone Deacetylase Functions in Gastric Cancer: Therapeutic Target?" Cancers 14, no. 21: 5472. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14215472