An Osteosarcoma Model by 3D Printed Polyurethane Scaffold and In Vitro Generated Bone Extracellular Matrix
Abstract
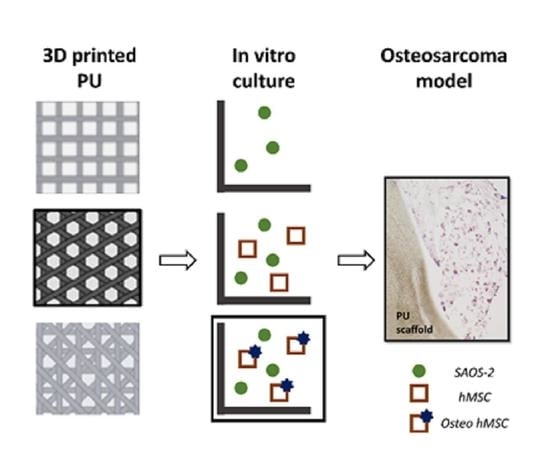
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. 3D Printing of PU Scaffolds
2.3. Morphological, Physical, and Mechanical Characterization
2.4. In Vitro Cytotoxicity Tests on PU Scaffolds
2.5. 3D Bone Model Preparation
2.6. 3D Osteosarcoma Model Preparation and Validation
2.7. Statistical Analysis
3. Results
3.1. Morphological, Physical, and Mechanical Properties of 3D Printed Polyurethane Scaffolds
3.2. In Vitro Cytotoxicity of PU Scaffolds
3.3. 3D In Vitro Bone Model
3.4. 3D In Vitro Osteosarcoma Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Caddeo, S.; Boffito, M.; Sartori, S. Tissue Engineering Approaches in the Design of Healthy and Pathological in Vitro Tissue Models. Front. Bioeng. Biotechnol. 2017, 5, 40. [Google Scholar] [CrossRef] [Green Version]
- Negrini, N.C.; Volponi, A.A.; Higgins, C.A.; Sharpe, P.T.; Celiz, A.D. Scaffold-Based Developmental Tissue Engineering Strategies for Ectodermal Organ Regeneration. Mater. Today Bio 2021, 10, 100107. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional Culture System of Cancer Cells Combined with Biomaterials for Drug Screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef]
- Bregenzer, M.E.; Horst, E.N.; Mehta, P.; Novak, C.M.; Raghavan, S.; Snyder, C.S.; Mehta, G. Integrated Cancer Tissue Engineering Models for Precision Medicine. PLoS ONE 2019, 14, e0216564. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Tang, Z.; Zhao, Y.; Yao, R.; Li, L. Three-Dimensional in Vitro Cancer Models: A Short Review. Biofabrication 2014, 14, 02201. [Google Scholar] [CrossRef]
- Ricci, C.; Moroni, L.; Danti, S. Cancer Tissue Engineering-New Perspectives in Understanding the Biology of Solid Tumours-a Critical Review. OA Tissue Eng. 2013, 1, 4. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.M.; Kolb, A.D.; Shupp, A.B.; Shine, K.M.; Bussard, K.M. Printing the Pathway Forward in Bone Metastatic Cancer Research: Applications of 3D Engineered Models and Bioprinted Scaffolds to Recapitulate the Bone—Tumor Niche. Cancers 2021, 13, 507. [Google Scholar] [CrossRef]
- Ricci, C.; Mota, C.; Moscato, S.; D’Alessandro, D.; Ugel, S.; Sartoris, S.; Bronte, V.; Boggi, U.; Campani, D.; Funel, N.; et al. Interfacing Polymeric Scaffolds with Primary Pancreatic Ductal Adenocarcinoma Cells to Develop 3D Cancer Models. Biomatter 2014, 4, e955386. [Google Scholar] [CrossRef] [Green Version]
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatol. Ther. 2017, 4, 25–43. [Google Scholar] [CrossRef] [Green Version]
- Sadykova, L.R.; Ntekim, A.I.; Muyangwa-Semenova, M.; Rutland, C.S.; Jeyapalan, J.N.; Blatt, N.; Rizvanov, A.A. Epidemiology and Risk Factors of Osteosarcoma. Cancer Investig. 2020, 38, 259–269. [Google Scholar] [CrossRef]
- Harrison, D.J.; Geller, D.S.; Gill, J.D.; Lewis, V.O.; Gorlick, R. Current and Future Therapeutic Approaches for Osteosarcoma. Expert Rev. Anticancer Ther. 2018, 18, 39–50. [Google Scholar] [CrossRef]
- Meazza, C.; Scanagatta, P. Metastatic Osteosarcoma: A Challenging Multidisciplinary Treatment. Expert Rev. Anticancer Ther. 2016, 16, 543–556. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)Evolution: Unveiling Tumor–Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef]
- Narkhede, A.A.; Crenshaw, J.H.; Crossman, D.K.; Shevde, L.A.; Rao, S.S. An in Vitro Hyaluronic Acid Hydrogel Based Platform to Model Dormancy in Brain Metastatic Breast Cancer Cells. Acta Biomater. 2020, 107, 65–77. [Google Scholar] [CrossRef]
- Lee, H.J.; Mun, S.; Pham, D.M.; Kim, P. Extracellular Matrix-Based Hydrogels to Tailoring Tumor Organoids. ACS Biomater. Sci. Eng. 2021, 7, 4128–4135. [Google Scholar] [CrossRef]
- Angeloni, V.; Contessi, N.; de Marco, C.; Bertoldi, S.; Tanzi, M.C.; Daidone, M.G.; Farè, S. Polyurethane Foam Scaffold as in Vitro Model for Breast Cancer Bone Metastasis. Acta Biomater. 2017, 63, 306–316. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, M.; Fu, Y.; Castro, N.J.; Fu, S.W.; Zhang, L.G. Engineering a Biomimetic Three-Dimensional Nanostructured Bone Model for Breast Cancer Bone Metastasis Study. Acta Biomater. 2015, 14, 164–174. [Google Scholar] [CrossRef]
- Talukdar, S.; Kundu, S.C. Engineered 3D Silk-Based Metastasis Models: Interactions between Human Breast Adenocarcinoma, Mesenchymal Stem Cells and Osteoblast-like Cells. Adv. Funct. Mater. 2013, 23, 5249–5260. [Google Scholar] [CrossRef]
- Subia, B.; Dey, T.; Sharma, S.; Kundu, S.C. Target Specific Delivery of Anticancer Drug in Silk Fibroin Based 3D Distribution Model of Bone-Breast Cancer Cells. ACS Appl. Mater. Interfaces 2015, 7, 2269–2279. [Google Scholar] [CrossRef]
- Pathi, S.P.; Kowalczewski, C.; Tadipatri, R.; Fischbach, C. A Novel 3-D Mineralized Tumor Model to Study Breast Cancer Bone Metastasis. PLoS ONE 2010, 5, e8849. [Google Scholar] [CrossRef]
- Pan, T.; Fong, E.L.S.; Martinez, M.; Harrington, D.A.; Lin, S.H.; Farach-Carson, M.C.; Satcher, R.L. Three-Dimensional (3D) Culture of Bone-Derived Human 786-O Renal Cell Carcinoma Retains Relevant Clinical Characteristics of Bone Metastases. Cancer Lett. 2015, 365, 89–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieh, S.; Taubenberger, A.V.; Lehman, M.L.; Clements, J.A.; Nelson, C.C.; Hutmacher, D.W. Paracrine Interactions between LNCaP Prostate Cancer Cells and Bioengineered Bone in 3D in Vitro Culture Reflect Molecular Changes during Bone Metastasis. Bone 2014, 63, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Lamhamedi-Cherradi, S.E.; Menegaz, B.A.; Ludwig, J.A.; Mikos, A.G. Flow Perfusion Effects on Three-Dimensional Culture and Drug Sensitivity of Ewing Sarcoma. Proc. Natl. Acad. Sci. USA 2015, 112, 10304–10309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, E.L.S.; Lamhamedi-Cherradi, S.E.; Burdett, E.; Ramamoorthy, V.; Lazar, A.J.; Kasper, F.K.; Farach-Carson, M.C.; Vishwamitra, D.; Demicco, E.G.; Menegaz, B.A.; et al. Modeling Ewing Sarcoma Tumors in Vitro with 3D Scaffolds. Proc. Natl. Acad. Sci. USA 2013, 110, 6500–6505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, P.H.S.; Aung, K.Z.; Toh, S.L.; Goh, J.C.H.; Nathan, S.S. Three-Dimensional Porous Silk Tumor Constructs in the Approximation of in Vivo Osteosarcoma Physiology. Biomaterials 2011, 32, 6131–6137. [Google Scholar] [CrossRef]
- Monteiro, C.F.; Custódio, C.A.; Mano, J.F. Bioengineering a Humanized 3D Tri-Culture Osteosarcoma Model to Assess Tumor Invasiveness and Therapy Response. Acta Biomater. 2021, 134, 204–214. [Google Scholar] [CrossRef]
- Costard, L.S.; Hosn, R.R.; Ramanayake, H.; O’Brien, F.J.; Curtin, C.M. Influences of the 3D Microenvironment on Cancer Cell Behaviour and Treatment Responsiveness: A Recent Update on Lung, Breast and Prostate Cancer Models. Acta Biomater. 2021, 132, 360–378. [Google Scholar] [CrossRef]
- Griffin, M.; Castro, N.; Bas, O.; Saifzadeh, S.; Butler, P.; Hutmacher, D.W. The Current Versatility of Polyurethane Three-Dimensional Printing for Biomedical Applications. Tissue Eng. Part B Rev. 2020, 26, 272–283. [Google Scholar] [CrossRef]
- Datta, N.; Holtorf, H.L.; Sikavitsas, V.I.; Jansen, J.A.; Mikos, A.G. Effect of Bone Extracellular Matrix Synthesized in Vitro on the Osteoblastic Differentiation of Marrow Stromal Cells. Biomaterials 2005, 26, 971–977. [Google Scholar] [CrossRef]
- Datta, N.; Pham, Q.P.; Sharma, U.; Sikavitsas, V.I.; Jansen, J.A.; Mikos, A.G. In Vitro Generated Extracellular Matrix and Fluid Shear Stress Synergistically Enhance 3D Osteoblastic Differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 2488–2493. [Google Scholar] [CrossRef] [Green Version]
- Danti, S.; Stefanini, C.; D’Alessandro, D.; Moscato, S.; Pietrabissa, A.; Petrini, M.; Berrettini, S. Novel Biological/Biohybrid Prostheses for the Ossicular Chain: Fabrication Feasibility and Preliminary Functional Characterization. Biomed. Microdevices 2009, 11, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Campiglio, C.E.; Ponzini, S.; de Stefano, P.; Ortoleva, G.; Vignati, L.; Draghi, L. Cross-Linking Optimization for Electrospun Gelatin: Challenge of Preserving Fiber Topography. Polymers 2020, 12, 2472. [Google Scholar] [CrossRef] [PubMed]
- Negrini, N.C.; Celikkin, N.; Tarsini, P.; Farè, S.; Święszkowski, W. Three-Dimensional Printing of Chemically Crosslinked Gelatin Hydrogels for Adipose Tissue Engineering. Biofabrication 2020, 12, 025001. [Google Scholar] [CrossRef] [PubMed]
- Agassant, J.F.; Pigeonneau, F.; Sardo, L.; Vincent, M. Flow Analysis of the Polymer Spreading during Extrusion Additive Manufacturing. Addit. Manuf. 2019, 29, 100794. [Google Scholar] [CrossRef] [Green Version]
- Betriu, N.; Andreeva, A.; Semino, C.E. Erlotinib Promotes Ligand-Induced EGFR Degradation in 3D but Not 2D Cultures of Pancreatic Ductal Adenocarcinoma Cells. Cancers 2021, 13, 4504. [Google Scholar] [CrossRef]
- Fischetti, T.; Di Pompo, G.; Baldini, N.; Avnet, S.; Graziani, G. 3d Printing and Bioprinting to Model Bone Cancer: The Role of Materials and Nanoscale Cues in Directing Cell Behavior. Cancers 2021, 13, 4065. [Google Scholar] [CrossRef]
- Milazzo, M.; Contessi Negrini, N.; Scialla, S.; Marelli, B.; Farè, S.; Danti, S.; Buehler, M.J. Additive Manufacturing Approaches for Hydroxyapatite-Reinforced Composites. Adv. Funct. Mater. 2019, 29, 1903055. [Google Scholar] [CrossRef] [Green Version]
- Meskinfam, M.; Bertoldi, S.; Albanese, N.; Cerri, A.; Tanzi, M.C.; Imani, R.; Baheiraei, N.; Farokhi, M.; Farè, S. Polyurethane Foam/Nano Hydroxyapatite Composite as a Suitable Scaffold for Bone Tissue Regeneration. Mater. Sci. Eng. C 2018, 82, 130–140. [Google Scholar] [CrossRef]
- Pitton, M.; Fiorati, A.; Buscemi, S.; Melone, L.; Farè, S.; Contessi Negrini, N. 3D Bioprinting of Pectin-Cellulose Nanofibers Multicomponent Bioinks. Front. Bioeng. Biotechnol. 2021, 9, 732689. [Google Scholar] [CrossRef]
- Moroni, L.; de Wijn, J.R.; van Blitterswijk, C.A. 3D Fiber-Deposited Scaffolds for Tissue Engineering: Influence of Pores Geometry and Architecture on Dynamic Mechanical Properties. Biomaterials 2006, 27, 974–985. [Google Scholar] [CrossRef]
- Trachtenberg, J.E.; Santoro, M.; Williams, C.; Piard, C.M.; Smith, B.T.; Placone, J.K.; Menegaz, B.A.; Molina, E.R.; Lamhamedi-Cherradi, S.E.; Ludwig, J.A.; et al. Effects of Shear Stress Gradients on Ewing Sarcoma Cells Using 3D Printed Scaffolds and Flow Perfusion. ACS Biomater. Sci. Eng. 2018, 4, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Domingos, M.; Chiellini, F.; Cometa, S.; de Giglio, E.; Grillo-Fernandes, E.; Bártolo, P.; Chiellini, E. Evaluation of in Vitro Degradation of Pcl Scaffolds Fabricated via Bioextrusion. Part 1: Influence of the Degradation Environment. Virtual Phys. Prototyp. 2010, 5, 65–73. [Google Scholar] [CrossRef]
- Lynch, M.E.; Chiou, A.E.; Lee, M.J.; Marcott, S.C.; Polamraju, P.V.; Lee, Y.; Fischbach, C. Three-Dimensional Mechanical Loading Modulates the Osteogenic Response of Mesenchymal Stem Cells to Tumor-Derived Soluble Signals. Tissue Eng. Part A 2016, 22, 1006–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Pastakia, M.; Fenn, M.B.; Kishore, V. Saos-2 Cell-Mediated Mineralization on Collagen Gels: Effect of Densification and Bioglass Incorporation. J. Biomed. Mater. Res. Part A 2016, 104, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Contessi Negrini, N.; Angelova Volponi, A.; Sharpe, P.T.; Celiz, A.D. Tunable Cross-Linking and Adhesion of Gelatin Hydrogels via Bioorthogonal Click Chemistry. ACS Biomater. Sci. Eng. 2021, 7, 4330–4346. [Google Scholar] [CrossRef]
- Kostic, A.; Lynch, C.D.; Sheetz, M.P. Differential Matrix Rigidity Response in Breast Cancer Cell Lines Correlates with the Tissue Tropism. PLoS ONE 2009, 4, e6361. [Google Scholar] [CrossRef]
- Al-Munajjed, A.A.; Plunkett, N.A.; Gleeson, J.P.; Weber, T.; Jungreuthmayer, C.; Levingstone, T.; Hammer, J.; O’Brien, F.J. Development of a Biomimetic Collagen-Hydroxyapatite Scaffold for Bone Tissue Engineering Using a SBF Immersion Technique. J. Biomed. Mater. Res. Part B Appl. Biomater. 2009, 90, 584–591. [Google Scholar] [CrossRef]
- Zhu, W.; Castro, N.J.; Cui, H.; Zhou, X.; Boualam, B.; McGrane, R.; Glazer, R.I.; Zhang, L.G. A 3D Printed Nano Bone Matrix for Characterization of Breast Cancer Cell and Osteoblast Interactions. Nanotechnology 2016, 27, 315103. [Google Scholar] [CrossRef]
- Miao, H.; Shen, R.; Zhang, W.; Lin, Z.; Wang, H.; Yang, L.; Liu, X.Y.; Lin, N. Near-Infrared Light Triggered Silk Fibroin Scaffold for Photothermal Therapy and Tissue Repair of Bone Tumors. Adv. Funct. Mater. 2021, 31, 2007188. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor Microenvironment Complexity and Therapeutic Implications at a Glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [Green Version]
- Lorusso, G.; Rüegg, C.; Kuonen, F. Targeting the Extra-Cellular Matrix—Tumor Cell Crosstalk for Anti-Cancer Therapy: Emerging Alternatives to Integrin Inhibitors. Front. Oncol. 2020, 10, 01231. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, L.; Cascone, M.G.; Danti, S.; Serino, L.P.; Moscato, S.; Bernardini, N. Gelatine/PLLA Sponge-like Scaffolds: Morphological and Biological Characterization. J. Mater. Sci. Mater. Med. 2007, 18, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Moscato, S.; Ronca, F.; Campani, D.; Danti, S. Poly(Vinyl Alcohol)/Gelatin Hydrogels Cultured with HepG2 Cells as a 3D Model of Hepatocellular Carcinoma: A Morphological Study. J. Funct. Biomater. 2015, 6, 16–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, V. TGF-β Signaling in Cancer. J. Cell. Biochem. 2016, 117, 1279–1287. [Google Scholar] [CrossRef]
- Liu, J.; Wu, J.; Zhou, L.; Pan, C.; Zhou, Y.; Du, W.; Chen, J.-M.; Zhu, X.; Shen, J.; Chen, S.; et al. ZD6474, a New Treatment Strategy for Human Osteosarcoma, and Its Potential Synergistic Effect with Celecoxib. Oncotarget 2015, 6, 21341. [Google Scholar] [CrossRef]
- Viti, F.; Landini, M.; Mezzelani, A.; Petecchia, L.; Milanesi, L.; Scaglione, S. Osteogenic Differentiation of MSC through Calcium Signaling Activation: Transcriptomics and Functional Analysis. PLoS ONE 2016, 11, e0148173. [Google Scholar] [CrossRef] [Green Version]
- de la Ossa, J.G.; Trombi, L.; D’Alessandro, D.; Coltelli, M.B.; Serino, L.P.; Pini, R.; Lazzeri, A.; Petrini, M.; Danti, S. Pore Size Distribution and Blend Composition Affect In Vitro Prevascularized Bone Matrix Formation on Poly(Vinyl Alcohol)/Gelatin Sponges. Macromol. Mater. Eng. 2017, 302, 1700300. [Google Scholar] [CrossRef]
- Grolman, J.M.; Zhang, D.; Smith, A.M.; Moore, J.S.; Kilian, K.A. Rapid 3D Extrusion of Synthetic Tumor Microenvironments. Adv. Mater. 2015, 27, 5512–5517. [Google Scholar] [CrossRef] [Green Version]









| Filament Distance [mm] | Angle Shift [°] | ||
|---|---|---|---|
| 90° | 60° | 45° | |
| 0.7 | 90_0.7 | 60_0.7 | 45_0.7 |
| 1.0 | 90_1 | 60_1 | 45_1 |
| 3D Osteosarcoma Model Acronym | Scaffold Pre-Treatment (1st Step Culture, 3 Weeks) | Substrate Used for Osteosarcoma Cells | 2nd Step Culture (1 Week) |
|---|---|---|---|
| PU | None | Plain scaffold | SAOS-2 cells |
| PU_ECM | Undifferentiated hMSC culture followed by cell lysis | Scaffold containing immature bone ECM | SAOS-2 cells |
| PU_bECM | Osteo-differentiated hMSC culture followed by cell lysis | Scaffold containing mature (mineralized) bone ECM | SAOS-2 cells |
| Printing Parameters | Printing Speed [mm∙s−1] | Extrusion Temperature [°C] | Plate Temperature [°C] | Extrusion Multiplier | ||
| 12 | 240 | 30 | 1.5 | |||
| Design parameters | Layer height [mm] | Filament thickness [mm] | Filament diameter [mm] | |||
| 0.4 | 0.4 | 1.7 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contessi Negrini, N.; Ricci, C.; Bongiorni, F.; Trombi, L.; D’Alessandro, D.; Danti, S.; Farè, S. An Osteosarcoma Model by 3D Printed Polyurethane Scaffold and In Vitro Generated Bone Extracellular Matrix. Cancers 2022, 14, 2003. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14082003
Contessi Negrini N, Ricci C, Bongiorni F, Trombi L, D’Alessandro D, Danti S, Farè S. An Osteosarcoma Model by 3D Printed Polyurethane Scaffold and In Vitro Generated Bone Extracellular Matrix. Cancers. 2022; 14(8):2003. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14082003
Chicago/Turabian StyleContessi Negrini, Nicola, Claudio Ricci, Federica Bongiorni, Luisa Trombi, Delfo D’Alessandro, Serena Danti, and Silvia Farè. 2022. "An Osteosarcoma Model by 3D Printed Polyurethane Scaffold and In Vitro Generated Bone Extracellular Matrix" Cancers 14, no. 8: 2003. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers14082003